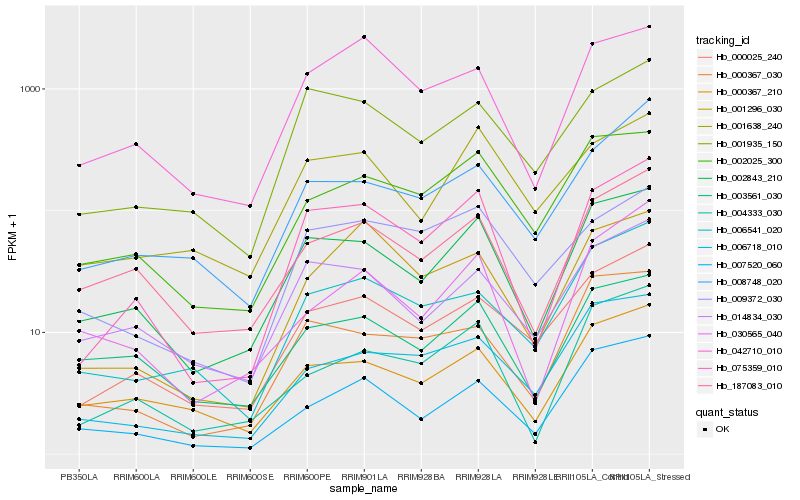
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075359_010 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_002843_210 |
0.1060159709 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 3 |
Hb_001935_150 |
0.1347358526 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 4 |
Hb_042710_010 |
0.1367044215 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 5 |
Hb_000025_240 |
0.1377633925 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 6 |
Hb_004333_030 |
0.1402734973 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 7 |
Hb_187083_010 |
0.1454055835 |
- |
- |
PREDICTED: uncharacterized protein LOC105643971 [Jatropha curcas] |
| 8 |
Hb_001296_030 |
0.1455822129 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |
| 9 |
Hb_009372_030 |
0.1495340903 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 10 |
Hb_014834_030 |
0.1526784764 |
- |
- |
- |
| 11 |
Hb_001638_240 |
0.1527319853 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 12 |
Hb_000367_030 |
0.1538970431 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 13 |
Hb_002025_300 |
0.1539648727 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 14 |
Hb_008748_020 |
0.1584242121 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 15 |
Hb_006541_020 |
0.158809596 |
- |
- |
unknown [Lotus japonicus] |
| 16 |
Hb_000367_210 |
0.1591609374 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 17 |
Hb_007520_060 |
0.1593709257 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 18 |
Hb_006718_010 |
0.1657565566 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 19 |
Hb_003561_030 |
0.1673034045 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |
| 20 |
Hb_030565_040 |
0.1683443333 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |