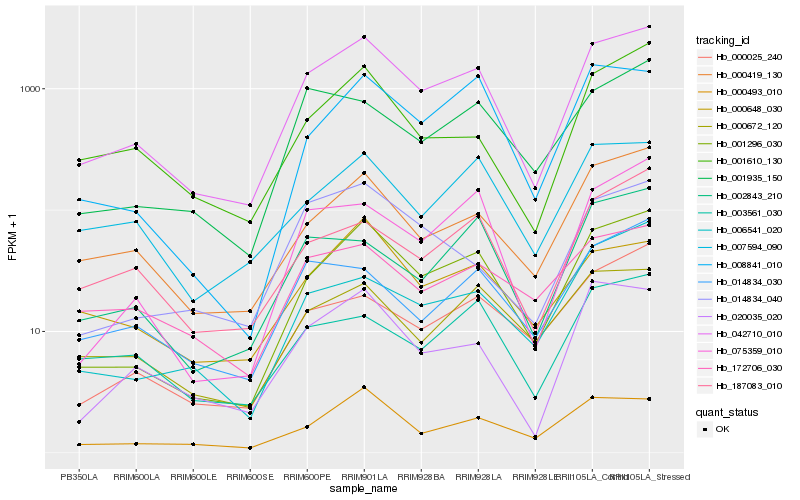
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042710_010 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 2 |
Hb_001296_030 |
0.0860404228 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |
| 3 |
Hb_000419_130 |
0.1134852304 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |
| 4 |
Hb_020035_020 |
0.1224377169 |
- |
- |
PREDICTED: uncharacterized protein LOC105631020 [Jatropha curcas] |
| 5 |
Hb_000493_010 |
0.1269325149 |
- |
- |
hypothetical protein CICLE_v10021431mg [Citrus clementina] |
| 6 |
Hb_007594_090 |
0.1338619756 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18 [Jatropha curcas] |
| 7 |
Hb_001610_130 |
0.1345697271 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 8 |
Hb_075359_010 |
0.1367044215 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 9 |
Hb_187083_010 |
0.1370323926 |
- |
- |
PREDICTED: uncharacterized protein LOC105643971 [Jatropha curcas] |
| 10 |
Hb_000672_120 |
0.1378653551 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 11 |
Hb_000648_030 |
0.1382579331 |
- |
- |
hypothetical protein PHAVU_002G055100g [Phaseolus vulgaris] |
| 12 |
Hb_001935_150 |
0.1385948504 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 13 |
Hb_014834_040 |
0.1419366709 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 14 |
Hb_008841_010 |
0.1423062168 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 15 |
Hb_172706_030 |
0.1430332898 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 16 |
Hb_002843_210 |
0.1433362374 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 17 |
Hb_014834_030 |
0.1437787487 |
- |
- |
- |
| 18 |
Hb_003561_030 |
0.1459644991 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |
| 19 |
Hb_000025_240 |
0.1490738753 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 20 |
Hb_006541_020 |
0.1512635851 |
- |
- |
unknown [Lotus japonicus] |