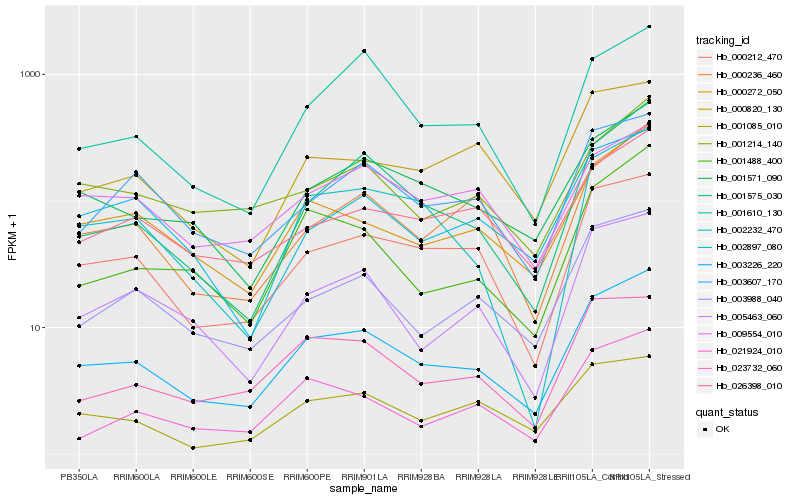
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_220 |
0.0 |
- |
- |
hypothetical protein CICLE_v10022836mg [Citrus clementina] |
| 2 |
Hb_000212_470 |
0.0923210073 |
- |
- |
hypothetical protein CICLE_v10033256mg [Citrus clementina] |
| 3 |
Hb_001571_090 |
0.0990691152 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 4 |
Hb_000272_050 |
0.1023358013 |
- |
- |
hypothetical protein JCGZ_03991 [Jatropha curcas] |
| 5 |
Hb_003988_040 |
0.1029955754 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 6 |
Hb_001575_030 |
0.1078854689 |
- |
- |
hypothetical protein POPTR_0022s00880g [Populus trichocarpa] |
| 7 |
Hb_005463_060 |
0.1091537762 |
- |
- |
PREDICTED: uncharacterized protein LOC105632515 [Jatropha curcas] |
| 8 |
Hb_001488_400 |
0.1148165114 |
- |
- |
hypothetical protein POPTR_0018s12030g [Populus trichocarpa] |
| 9 |
Hb_002232_470 |
0.11513561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000236_460 |
0.116011855 |
- |
- |
PREDICTED: uncharacterized protein LOC105633119 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003607_170 |
0.1161174555 |
- |
- |
U6 snRNA-associated Sm-like protein LSm5 [Hevea brasiliensis] |
| 12 |
Hb_026398_010 |
0.1177480083 |
- |
- |
hypothetical protein B456_010G132100 [Gossypium raimondii] |
| 13 |
Hb_023732_060 |
0.1180142085 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |
| 14 |
Hb_001085_010 |
0.1195684183 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 4 isoform X3 [Populus euphratica] |
| 15 |
Hb_000820_130 |
0.1223291398 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 16 |
Hb_002897_080 |
0.1226894993 |
- |
- |
hypothetical protein JCGZ_03306 [Jatropha curcas] |
| 17 |
Hb_001610_130 |
0.1264113826 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 18 |
Hb_001214_140 |
0.1270806844 |
- |
- |
60S ribosomal protein L27a, putative [Ricinus communis] |
| 19 |
Hb_021924_010 |
0.1278952066 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 20 |
Hb_009554_010 |
0.1293424827 |
- |
- |
PREDICTED: multifunctional methyltransferase subunit TRM112-like protein At1g22270 [Jatropha curcas] |