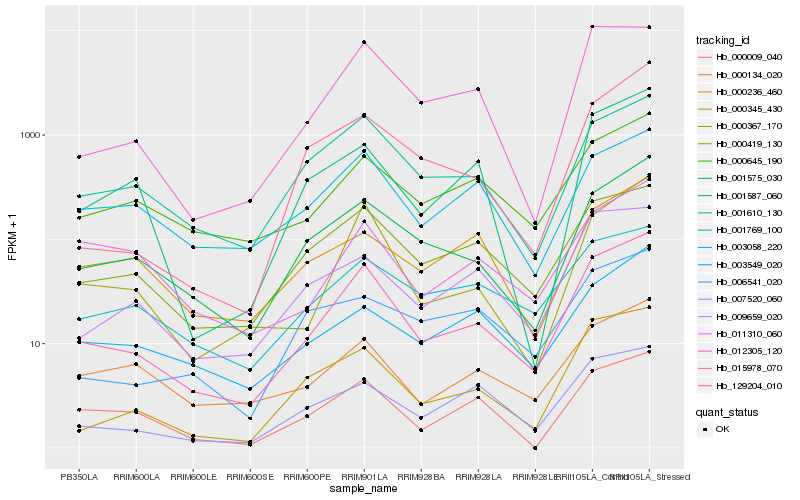
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015978_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000367_170 |
0.1152560483 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_001575_030 |
0.1180047113 |
- |
- |
hypothetical protein POPTR_0022s00880g [Populus trichocarpa] |
| 4 |
Hb_000345_430 |
0.1226671884 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 5 |
Hb_001610_130 |
0.1285259314 |
- |
- |
40S ribosomal protein S30 [Medicago truncatula] |
| 6 |
Hb_012305_120 |
0.1334501629 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 7 |
Hb_000419_130 |
0.1343560394 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |
| 8 |
Hb_007520_060 |
0.1429471884 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 9 |
Hb_000645_190 |
0.1475231369 |
- |
- |
PREDICTED: 60S ribosomal protein L29-1-like [Glycine max] |
| 10 |
Hb_009659_020 |
0.1502809089 |
- |
- |
PREDICTED: uncharacterized protein LOC103870090 [Brassica rapa] |
| 11 |
Hb_001587_060 |
0.1537977127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000009_040 |
0.1560539003 |
- |
- |
- |
| 13 |
Hb_001769_100 |
0.1582059924 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 14 |
Hb_000134_020 |
0.1592397201 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 15 |
Hb_129204_010 |
0.1594899905 |
- |
- |
LOC100284182 [Zea mays] |
| 16 |
Hb_003549_020 |
0.1615464718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003058_220 |
0.1638292396 |
- |
- |
Pre-mRNA branch site protein p14, putative [Ricinus communis] |
| 18 |
Hb_006541_020 |
0.1728716492 |
- |
- |
unknown [Lotus japonicus] |
| 19 |
Hb_011310_060 |
0.1739573033 |
- |
- |
PREDICTED: uncharacterized protein At2g34160-like [Jatropha curcas] |
| 20 |
Hb_000236_460 |
0.1745462753 |
- |
- |
PREDICTED: uncharacterized protein LOC105633119 isoform X1 [Jatropha curcas] |