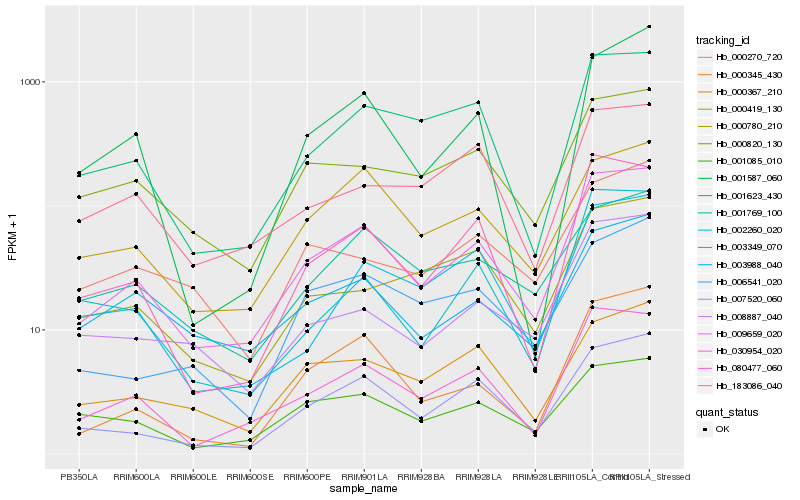
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009659_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103870090 [Brassica rapa] |
| 2 |
Hb_030954_020 |
0.0757082162 |
- |
- |
hypothetical protein RCOM_1330200 [Ricinus communis] |
| 3 |
Hb_007520_060 |
0.0956660471 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 4 |
Hb_080477_060 |
0.1097976893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000345_430 |
0.1146881148 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 6 |
Hb_001623_430 |
0.1219666635 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 7 |
Hb_000820_130 |
0.1233575639 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 8 |
Hb_003988_040 |
0.1245402628 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 9 |
Hb_008887_040 |
0.1261091102 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000419_130 |
0.1274157281 |
- |
- |
PREDICTED: uncharacterized protein LOC105650901 [Jatropha curcas] |
| 11 |
Hb_003349_070 |
0.1316564758 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 12 |
Hb_000367_210 |
0.1318766056 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 13 |
Hb_001769_100 |
0.1359811489 |
- |
- |
hypothetical protein POPTR_0014s01620g [Populus trichocarpa] |
| 14 |
Hb_006541_020 |
0.1373528544 |
- |
- |
unknown [Lotus japonicus] |
| 15 |
Hb_001085_010 |
0.1392207905 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 4 isoform X3 [Populus euphratica] |
| 16 |
Hb_000270_720 |
0.1393685381 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 17 |
Hb_002260_020 |
0.1401513301 |
- |
- |
- |
| 18 |
Hb_000780_210 |
0.1402258087 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 19 |
Hb_183086_040 |
0.1406309317 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 20 |
Hb_001587_060 |
0.1409742735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |