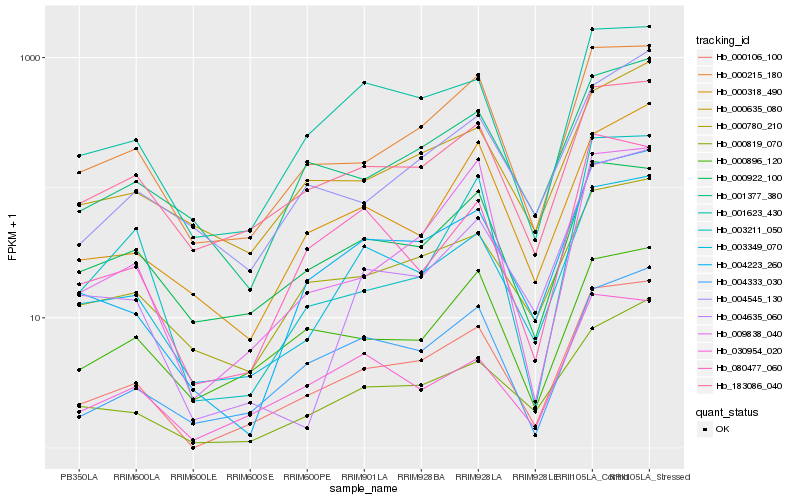
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_000215_180 |
0.0982730128 |
- |
- |
hypothetical protein 17 [Hevea brasiliensis] |
| 3 |
Hb_003349_070 |
0.1020763773 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Gossypium raimondii] |
| 4 |
Hb_004223_260 |
0.1052160056 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 5 |
Hb_000780_210 |
0.1164778016 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 6 |
Hb_001623_430 |
0.1166130025 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 7 |
Hb_183086_040 |
0.1167162042 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 8 |
Hb_000819_070 |
0.1226767422 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 9 |
Hb_004333_030 |
0.1249240023 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 10 |
Hb_030954_020 |
0.1309365539 |
- |
- |
hypothetical protein RCOM_1330200 [Ricinus communis] |
| 11 |
Hb_001377_380 |
0.1320486639 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 12 |
Hb_009838_040 |
0.1340551013 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 13 |
Hb_000922_100 |
0.1349978707 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 14 |
Hb_000635_080 |
0.1381794373 |
- |
- |
60S ribosomal protein L17A [Hevea brasiliensis] |
| 15 |
Hb_080477_060 |
0.1385786578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000318_490 |
0.1419570063 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM6 homolog [Jatropha curcas] |
| 17 |
Hb_000896_120 |
0.1455324059 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 18 |
Hb_003211_050 |
0.1516741794 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 19 |
Hb_004545_130 |
0.1521857468 |
- |
- |
uncharacterized protein LOC100305465 [Glycine max] |
| 20 |
Hb_004635_060 |
0.1525871393 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |