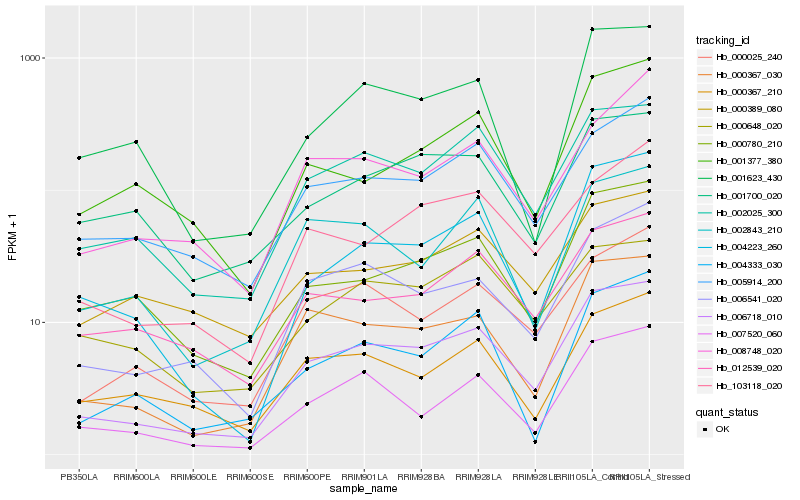
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006718_010 |
0.0 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 2 |
Hb_002025_300 |
0.0897870333 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 3 |
Hb_000367_030 |
0.0944817326 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 4 |
Hb_006541_020 |
0.099875494 |
- |
- |
unknown [Lotus japonicus] |
| 5 |
Hb_000025_240 |
0.1088866109 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 6 |
Hb_004223_260 |
0.1133044049 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 7 |
Hb_007520_060 |
0.1140374191 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 8 |
Hb_005914_200 |
0.1181446393 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 9 |
Hb_000780_210 |
0.1204510221 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 10 |
Hb_103118_020 |
0.1227739003 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 11 |
Hb_012539_020 |
0.128248607 |
- |
- |
protein binding/ubiquitin-protein ligase 4 [Hevea brasiliensis] |
| 12 |
Hb_001623_430 |
0.131838194 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 13 |
Hb_004333_030 |
0.1329822419 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 14 |
Hb_001377_380 |
0.136767999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 15 |
Hb_000367_210 |
0.1372143876 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 16 |
Hb_002843_210 |
0.1381394652 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 17 |
Hb_000648_020 |
0.1430827863 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 18 |
Hb_000389_080 |
0.1439279427 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 19 |
Hb_008748_020 |
0.1439663606 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |
| 20 |
Hb_001700_020 |
0.1453005807 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |