| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
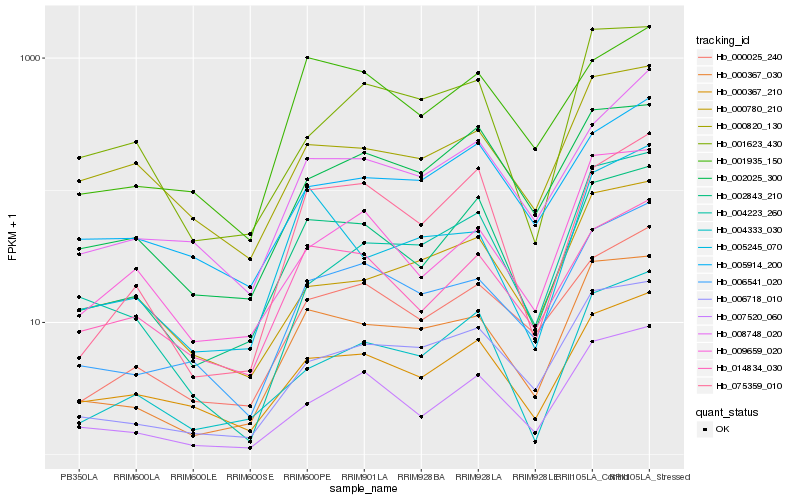
Hb_000367_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 2 |
Hb_006718_010 |
0.0944817326 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 3 |
Hb_006541_020 |
0.1117947027 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_002843_210 |
0.1236268307 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 5 |
Hb_005245_070 |
0.1287208331 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 6 |
Hb_000025_240 |
0.1351702412 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 7 |
Hb_007520_060 |
0.1369871298 |
- |
- |
PREDICTED: putative exosome complex component rrp40 [Jatropha curcas] |
| 8 |
Hb_002025_300 |
0.1407770629 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 9 |
Hb_000367_210 |
0.1416064667 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 10 |
Hb_001623_430 |
0.1493540239 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |
| 11 |
Hb_009659_020 |
0.1499487609 |
- |
- |
PREDICTED: uncharacterized protein LOC103870090 [Brassica rapa] |
| 12 |
Hb_000780_210 |
0.1505232244 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 13 |
Hb_004223_260 |
0.1530011042 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 14 |
Hb_075359_010 |
0.1538970431 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 15 |
Hb_004333_030 |
0.1544285458 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 16 |
Hb_000820_130 |
0.1551398137 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 17 |
Hb_014834_030 |
0.1555587653 |
- |
- |
- |
| 18 |
Hb_001935_150 |
0.1594947935 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 19 |
Hb_005914_200 |
0.1598579323 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 20 |
Hb_008748_020 |
0.1615815209 |
- |
- |
hypothetical protein B456_005G210700 [Gossypium raimondii] |