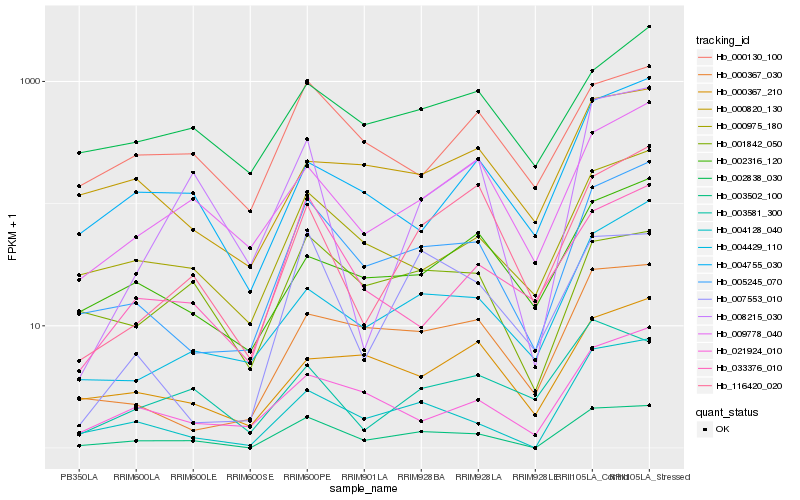
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003502_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_005245_070 |
0.1466588837 |
- |
- |
PREDICTED: uncharacterized protein LOC105643658 [Jatropha curcas] |
| 3 |
Hb_004128_040 |
0.1557751314 |
- |
- |
- |
| 4 |
Hb_000975_180 |
0.1824916359 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 5 |
Hb_001842_050 |
0.2012316292 |
- |
- |
PREDICTED: protein BUD31 homolog 1-like [Jatropha curcas] |
| 6 |
Hb_000367_030 |
0.2044706849 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 7 |
Hb_009778_040 |
0.205269881 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 8 |
Hb_021924_010 |
0.2057840498 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 9 |
Hb_008215_030 |
0.2104751274 |
- |
- |
hypothetical protein 3 [Hevea brasiliensis] |
| 10 |
Hb_000130_100 |
0.2135421995 |
- |
- |
hypothetical protein CICLE_v10022662mg [Citrus clementina] |
| 11 |
Hb_033376_010 |
0.2141747694 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 12 |
Hb_007553_010 |
0.2153295696 |
- |
- |
- |
| 13 |
Hb_116420_020 |
0.2176716931 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 14 |
Hb_002838_030 |
0.223699852 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 15 |
Hb_004429_110 |
0.2245621022 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 16 |
Hb_002316_120 |
0.224686176 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 17 |
Hb_000367_210 |
0.2255286653 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 18 |
Hb_000820_130 |
0.2259657877 |
- |
- |
PREDICTED: uncharacterized protein LOC105769309 isoform X1 [Gossypium raimondii] |
| 19 |
Hb_004755_030 |
0.2267708591 |
- |
- |
EG964 [Manihot glaziovii] |
| 20 |
Hb_003581_300 |
0.2278519617 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |