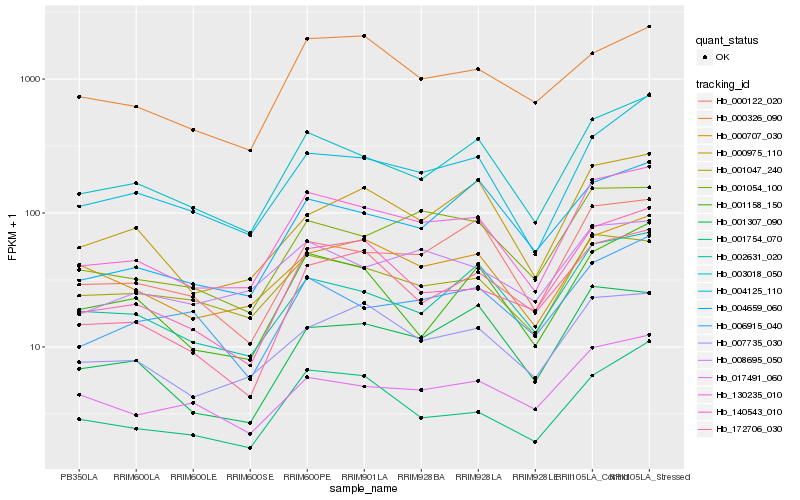
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_130235_010 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 2 |
Hb_003018_050 |
0.0769989596 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase Pin1 [Jatropha curcas] |
| 3 |
Hb_004125_110 |
0.0954513204 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 4 |
Hb_001054_100 |
0.0976716918 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 5 |
Hb_007735_030 |
0.0990932178 |
- |
- |
Calcium-binding EF-hand family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_140543_010 |
0.1007070939 |
- |
- |
- |
| 7 |
Hb_001754_070 |
0.1048811347 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 8 |
Hb_000122_020 |
0.1069380575 |
- |
- |
PREDICTED: alpha-soluble NSF attachment protein 2 [Jatropha curcas] |
| 9 |
Hb_172706_030 |
0.1081152624 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 10 |
Hb_002631_020 |
0.1084860803 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 11 |
Hb_008695_050 |
0.1090294872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001047_240 |
0.1142128873 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 20-like [Jatropha curcas] |
| 13 |
Hb_000707_030 |
0.114277373 |
- |
- |
PREDICTED: probable proteasome inhibitor [Vitis vinifera] |
| 14 |
Hb_000975_110 |
0.1157417915 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 [Jatropha curcas] |
| 15 |
Hb_001307_090 |
0.1158475586 |
- |
- |
hypothetical protein CISIN_1g0263261mg [Citrus sinensis] |
| 16 |
Hb_004659_060 |
0.1160253699 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 17 |
Hb_000326_090 |
0.1169071759 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 18 |
Hb_006915_040 |
0.1186129082 |
- |
- |
PREDICTED: prefoldin subunit 6 [Eucalyptus grandis] |
| 19 |
Hb_001158_150 |
0.1187834449 |
- |
- |
hypothetical protein JCGZ_26407 [Jatropha curcas] |
| 20 |
Hb_017491_060 |
0.1195963243 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |