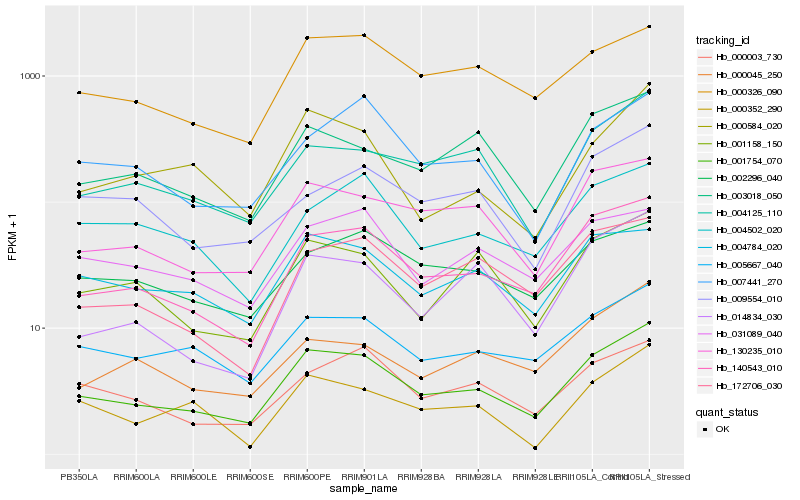
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 2 |
Hb_140543_010 |
0.0777165022 |
- |
- |
- |
| 3 |
Hb_130235_010 |
0.1048811347 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 4 |
Hb_005667_040 |
0.1107128463 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Jatropha curcas] |
| 5 |
Hb_001158_150 |
0.113009057 |
- |
- |
hypothetical protein JCGZ_26407 [Jatropha curcas] |
| 6 |
Hb_004125_110 |
0.1140960358 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 7 |
Hb_000326_090 |
0.1173330882 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 8 |
Hb_003018_050 |
0.1185195265 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase Pin1 [Jatropha curcas] |
| 9 |
Hb_000003_730 |
0.1191290594 |
- |
- |
DNA repair/transcription protein met18/mms19, putative [Ricinus communis] |
| 10 |
Hb_004784_020 |
0.1225496609 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 11 |
Hb_172706_030 |
0.122593613 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 12 |
Hb_014834_030 |
0.1277734306 |
- |
- |
- |
| 13 |
Hb_000584_020 |
0.1278526723 |
- |
- |
hypothetical protein PHAVU_002G233500g, partial [Phaseolus vulgaris] |
| 14 |
Hb_007441_270 |
0.1284132444 |
- |
- |
PREDICTED: 40S ribosomal protein S11 [Jatropha curcas] |
| 15 |
Hb_000045_250 |
0.1325320384 |
- |
- |
PREDICTED: uncharacterized protein LOC105646572 isoform X1 [Jatropha curcas] |
| 16 |
Hb_031089_040 |
0.1351299666 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |
| 17 |
Hb_004502_020 |
0.1351532066 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 18 |
Hb_002296_040 |
0.1352129748 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 19 |
Hb_000352_290 |
0.1355227004 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 20 |
Hb_009554_010 |
0.1358900512 |
- |
- |
PREDICTED: multifunctional methyltransferase subunit TRM112-like protein At1g22270 [Jatropha curcas] |