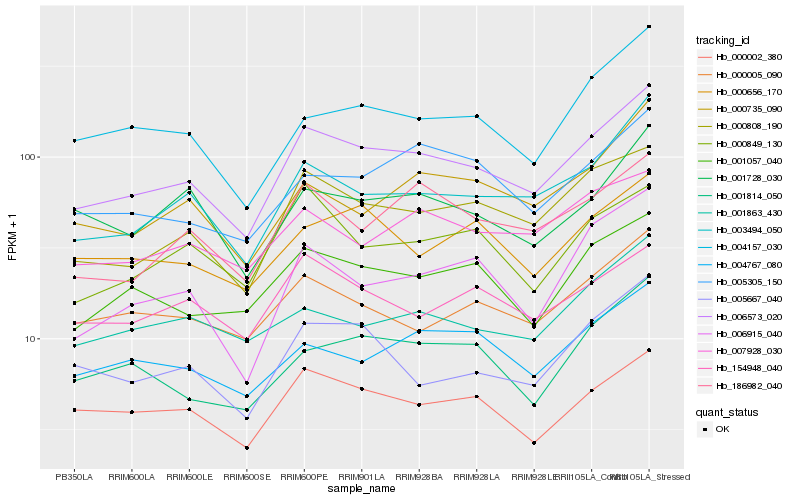
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006573_020 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S10-like [Jatropha curcas] |
| 2 |
Hb_000808_190 |
0.0664393405 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Eucalyptus grandis] |
| 3 |
Hb_003494_050 |
0.0756122428 |
- |
- |
hypothetical protein PRUPE_ppa013459mg [Prunus persica] |
| 4 |
Hb_001057_040 |
0.0830383361 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 5 |
Hb_000002_380 |
0.0844019299 |
- |
- |
PREDICTED: bet1-like SNARE 1-2 [Jatropha curcas] |
| 6 |
Hb_186982_040 |
0.0880558391 |
- |
- |
hypothetical protein PHAVU_003G089200g [Phaseolus vulgaris] |
| 7 |
Hb_000005_090 |
0.0909330155 |
- |
- |
hypothetical protein CICLE_v10029644mg [Citrus clementina] |
| 8 |
Hb_005667_040 |
0.0928088705 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 5 [Jatropha curcas] |
| 9 |
Hb_004157_030 |
0.0933261884 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 10 |
Hb_001814_050 |
0.0943333814 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_004767_080 |
0.0946311344 |
- |
- |
PREDICTED: mitochondrial ATPase complex subunit ATP10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007928_030 |
0.0949023784 |
- |
- |
PREDICTED: dual specificity phosphatase Cdc25 [Jatropha curcas] |
| 13 |
Hb_006915_040 |
0.0956598011 |
- |
- |
PREDICTED: prefoldin subunit 6 [Eucalyptus grandis] |
| 14 |
Hb_154948_040 |
0.0958455788 |
- |
- |
hypothetical protein B456_013G043800 [Gossypium raimondii] |
| 15 |
Hb_005305_150 |
0.0965658212 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 16 |
Hb_000735_090 |
0.0969605228 |
- |
- |
PREDICTED: 40S ribosomal protein S8-like [Gossypium raimondii] |
| 17 |
Hb_001728_030 |
0.0973724812 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 18 |
Hb_000849_130 |
0.0974926651 |
- |
- |
PREDICTED: adrenodoxin-like protein, mitochondrial [Nelumbo nucifera] |
| 19 |
Hb_001863_430 |
0.0976793627 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 20 |
Hb_000656_170 |
0.0978467296 |
- |
- |
lsm1, putative [Ricinus communis] |