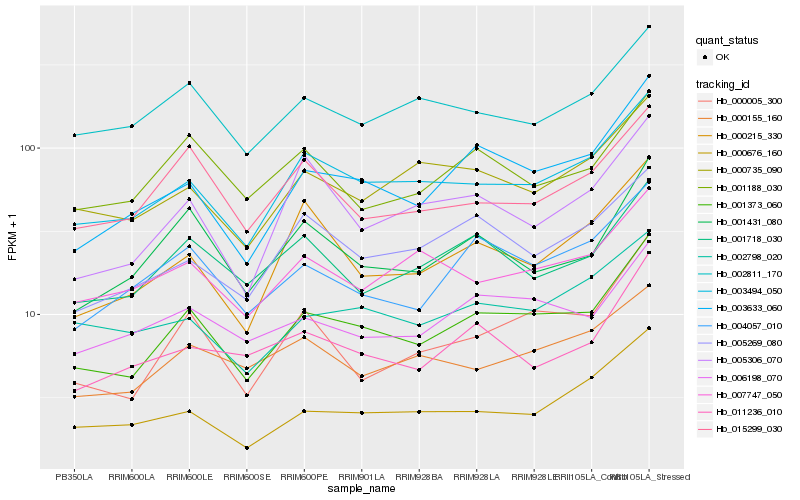
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001373_060 |
0.0 |
- |
- |
PREDICTED: structure-specific endonuclease subunit slx1 [Jatropha curcas] |
| 2 |
Hb_003633_060 |
0.07785358 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 3 |
Hb_003494_050 |
0.0781308997 |
- |
- |
hypothetical protein PRUPE_ppa013459mg [Prunus persica] |
| 4 |
Hb_004057_010 |
0.0952219191 |
- |
- |
PREDICTED: uncharacterized protein LOC105649313 [Jatropha curcas] |
| 5 |
Hb_000005_300 |
0.0965913335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006198_070 |
0.1028339838 |
- |
- |
pantoate-beta-alanine ligase, putative [Ricinus communis] |
| 7 |
Hb_001431_080 |
0.1029746822 |
- |
- |
40S ribosomal protein S11, putative [Ricinus communis] |
| 8 |
Hb_000735_090 |
0.1080792712 |
- |
- |
PREDICTED: 40S ribosomal protein S8-like [Gossypium raimondii] |
| 9 |
Hb_002798_020 |
0.1088319145 |
- |
- |
PREDICTED: uncharacterized protein LOC105647735 [Jatropha curcas] |
| 10 |
Hb_001718_030 |
0.1090479161 |
- |
- |
PREDICTED: calcineurin B-like protein 9 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000676_160 |
0.1092404075 |
- |
- |
folate carrier protein, putative [Ricinus communis] |
| 12 |
Hb_005269_080 |
0.1092604065 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_002811_170 |
0.114075535 |
- |
- |
PREDICTED: 40S ribosomal protein S3a-1 [Jatropha curcas] |
| 14 |
Hb_000215_330 |
0.1141895973 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 15 |
Hb_015299_030 |
0.1147568568 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 16 |
Hb_001188_030 |
0.1148835751 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 17 |
Hb_011236_010 |
0.114907287 |
- |
- |
hypothetical protein JCGZ_17006 [Jatropha curcas] |
| 18 |
Hb_000155_160 |
0.1155812766 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 19 |
Hb_005306_070 |
0.1156713703 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 20 |
Hb_007747_050 |
0.1160340963 |
- |
- |
Peptidyl-prolyl cis-trans isomerase cypE, putative [Ricinus communis] |