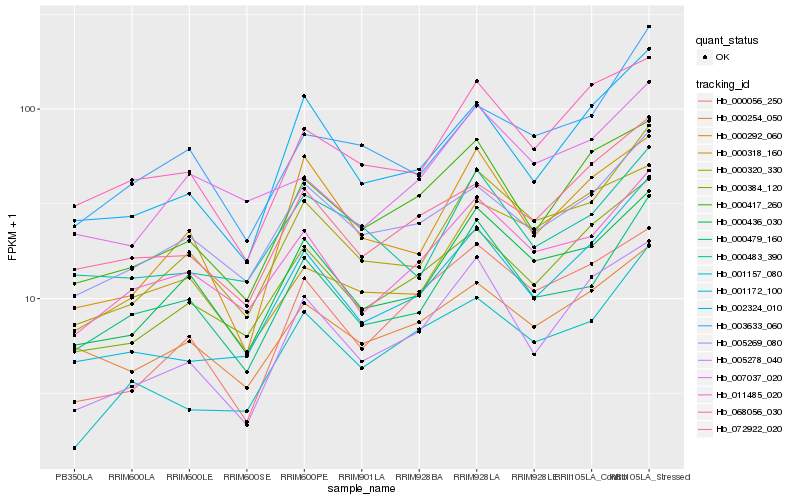
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_330 |
0.0 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 2 |
Hb_000436_030 |
0.0711158882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000384_120 |
0.0851027833 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 4 |
Hb_003633_060 |
0.0882513223 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 5 |
Hb_000479_160 |
0.08846665 |
- |
- |
Membrane-anchored ubiquitin-fold protein 1 precursor isoform 2 [Theobroma cacao] |
| 6 |
Hb_001172_100 |
0.0915951689 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_011485_020 |
0.0919587675 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_002324_010 |
0.0964924394 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 9 |
Hb_005269_080 |
0.0991928814 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_068056_030 |
0.1048532686 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 11 |
Hb_005278_040 |
0.1070963233 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |
| 12 |
Hb_000292_060 |
0.1089379782 |
- |
- |
transporter, putative [Ricinus communis] |
| 13 |
Hb_072922_020 |
0.1091484644 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |
| 14 |
Hb_000483_390 |
0.1143561234 |
- |
- |
PREDICTED: vesicle-associated protein 2-1 [Jatropha curcas] |
| 15 |
Hb_000417_260 |
0.1147928701 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000056_250 |
0.1149354964 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 17 |
Hb_000318_160 |
0.1153817716 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |
| 18 |
Hb_007037_020 |
0.1160490845 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 19 |
Hb_001157_080 |
0.1167677226 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000254_050 |
0.1169523769 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |