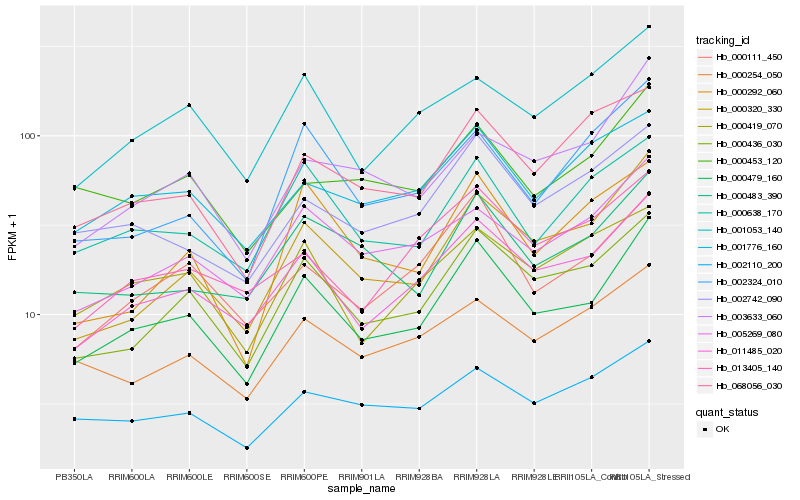
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_160 |
0.0 |
- |
- |
Membrane-anchored ubiquitin-fold protein 1 precursor isoform 2 [Theobroma cacao] |
| 2 |
Hb_011485_020 |
0.0723008218 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_000436_030 |
0.0780953165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000320_330 |
0.08846665 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 5 |
Hb_000111_450 |
0.0956793546 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 6 |
Hb_000483_390 |
0.10361625 |
- |
- |
PREDICTED: vesicle-associated protein 2-1 [Jatropha curcas] |
| 7 |
Hb_000638_170 |
0.1042827759 |
- |
- |
PREDICTED: ras-related protein RABC1 [Jatropha curcas] |
| 8 |
Hb_002742_090 |
0.104702331 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 9 |
Hb_005269_080 |
0.1055025902 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_001053_140 |
0.114072135 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001776_160 |
0.1141421331 |
- |
- |
20S proteasome alpha subunit G1 [Theobroma cacao] |
| 12 |
Hb_002110_200 |
0.1147494363 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_000254_050 |
0.1154880695 |
- |
- |
hypothetical protein VITISV_010510 [Vitis vinifera] |
| 14 |
Hb_068056_030 |
0.1170491425 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 15 |
Hb_000453_120 |
0.1179694458 |
- |
- |
PREDICTED: proteasome subunit alpha type-3 [Jatropha curcas] |
| 16 |
Hb_013405_140 |
0.117978466 |
- |
- |
PREDICTED: uncharacterized protein LOC105648437 [Jatropha curcas] |
| 17 |
Hb_002324_010 |
0.1182961882 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 18 |
Hb_000419_070 |
0.1190105383 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 19 |
Hb_003633_060 |
0.1196087124 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 20 |
Hb_000292_060 |
0.1200832662 |
- |
- |
transporter, putative [Ricinus communis] |