| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
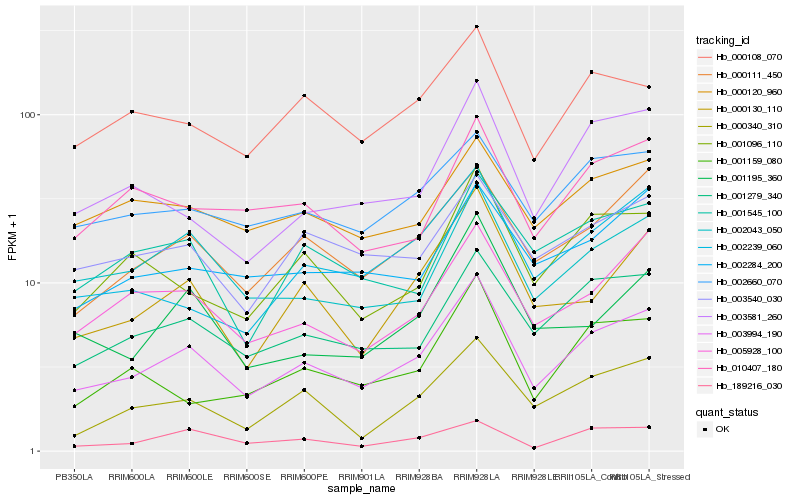
Hb_003994_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000111_450 |
0.0958062048 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 3 |
Hb_001279_340 |
0.0979477479 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 4 |
Hb_005928_100 |
0.1071071052 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2 [Jatropha curcas] |
| 5 |
Hb_000130_110 |
0.1088539584 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 6 |
Hb_000340_310 |
0.1120306672 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 7 |
Hb_002043_050 |
0.1121612632 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 8 |
Hb_002660_070 |
0.1174329873 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 9 |
Hb_001096_110 |
0.1188477995 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 10 |
Hb_003540_030 |
0.1192873924 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000108_070 |
0.1194400381 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 12 |
Hb_003581_260 |
0.1206423625 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 13 |
Hb_002239_060 |
0.1217991616 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 14 |
Hb_010407_180 |
0.1235622205 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 15 |
Hb_002284_200 |
0.1244954417 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001545_100 |
0.1248317558 |
- |
- |
PREDICTED: probable F-actin-capping protein subunit beta [Vitis vinifera] |
| 17 |
Hb_189216_030 |
0.1258173808 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 18 |
Hb_001195_360 |
0.1266517494 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 19 |
Hb_000120_960 |
0.1275092628 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_001159_080 |
0.1299995667 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |