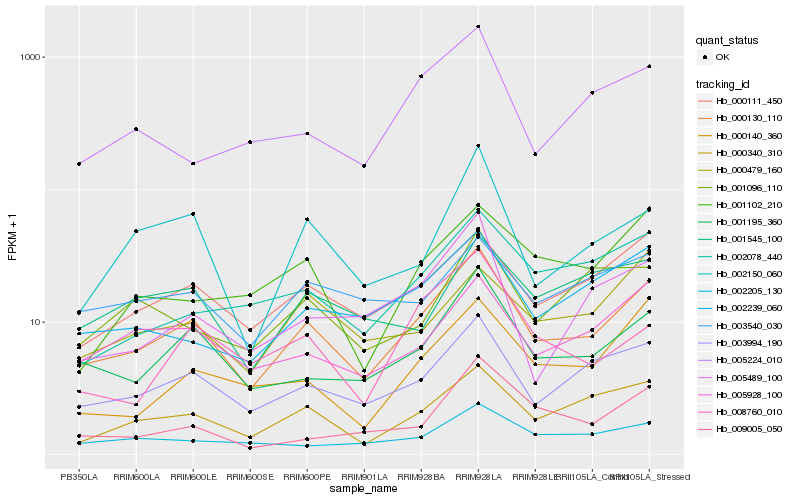
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_110 |
0.0 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 2 |
Hb_003994_190 |
0.1088539584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001195_360 |
0.1162463207 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 4 |
Hb_000340_310 |
0.1252602916 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 5 |
Hb_000111_450 |
0.1272397638 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 6 |
Hb_005928_100 |
0.1423144531 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2 [Jatropha curcas] |
| 7 |
Hb_002239_060 |
0.1443903405 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 8 |
Hb_002150_060 |
0.1533560667 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 9 |
Hb_002205_130 |
0.1566989625 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 10 |
Hb_002078_440 |
0.1586904664 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000140_360 |
0.1587058839 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 12 |
Hb_005489_100 |
0.1593862372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005224_010 |
0.1599790604 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 14 |
Hb_003540_030 |
0.1654745074 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001102_210 |
0.1689726796 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001545_100 |
0.1690846139 |
- |
- |
PREDICTED: probable F-actin-capping protein subunit beta [Vitis vinifera] |
| 17 |
Hb_001096_110 |
0.1701993147 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 18 |
Hb_008760_010 |
0.1705993662 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 19 |
Hb_009005_050 |
0.170886232 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 20 |
Hb_000479_160 |
0.1717040907 |
- |
- |
Membrane-anchored ubiquitin-fold protein 1 precursor isoform 2 [Theobroma cacao] |