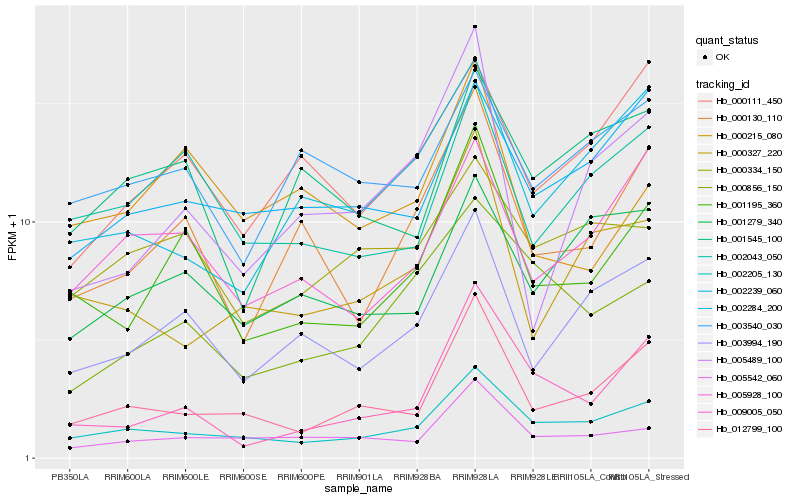
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_360 |
0.0 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 2 |
Hb_000130_110 |
0.1162463207 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 3 |
Hb_003994_190 |
0.1266517494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002043_050 |
0.1342076609 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 5 |
Hb_002205_130 |
0.1356715703 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 6 |
Hb_009005_050 |
0.1540056161 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 7 |
Hb_012799_100 |
0.1542239242 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 8 |
Hb_005928_100 |
0.1565148115 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2 [Jatropha curcas] |
| 9 |
Hb_005542_060 |
0.1605610617 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000215_080 |
0.1639620133 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000111_450 |
0.1749364362 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 12 |
Hb_005489_100 |
0.1753110068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000856_150 |
0.1786206386 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 14 |
Hb_002284_200 |
0.1826060828 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001279_340 |
0.1828783982 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 16 |
Hb_000327_220 |
0.1835205863 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 17 |
Hb_000334_150 |
0.1849250896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003540_030 |
0.1855198894 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001545_100 |
0.1856389293 |
- |
- |
PREDICTED: probable F-actin-capping protein subunit beta [Vitis vinifera] |
| 20 |
Hb_002239_060 |
0.1861100132 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |