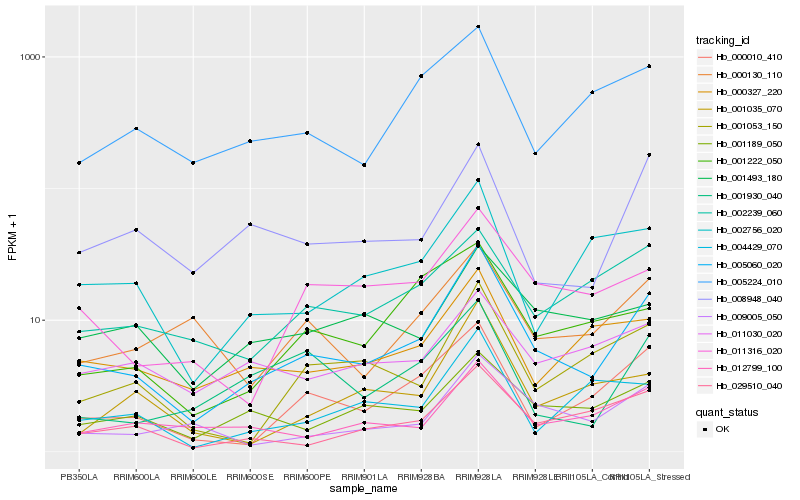
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005060_020 |
0.0 |
- |
- |
PREDICTED: metal tolerance protein 10-like isoform X1 [Populus euphratica] |
| 2 |
Hb_000010_410 |
0.163394882 |
- |
- |
hypothetical protein POPTR_0015s10070g, partial [Populus trichocarpa] |
| 3 |
Hb_029510_040 |
0.1772174201 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 4 |
Hb_009005_050 |
0.1809566374 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 5 |
Hb_001189_050 |
0.1818483204 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 6 |
Hb_001053_150 |
0.1823341844 |
- |
- |
- |
| 7 |
Hb_012799_100 |
0.1855123542 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 8 |
Hb_011316_020 |
0.1880750458 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 9 |
Hb_000327_220 |
0.1921109137 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 10 |
Hb_008948_040 |
0.1942676537 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 11 |
Hb_005224_010 |
0.1985110736 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 12 |
Hb_002756_020 |
0.1986986305 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 13 |
Hb_001035_070 |
0.1997293271 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 14 |
Hb_002239_060 |
0.2004965305 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 15 |
Hb_001222_050 |
0.2011892236 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |
| 16 |
Hb_001930_040 |
0.2025456927 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_001493_180 |
0.2047652155 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 18 |
Hb_000130_110 |
0.2071619615 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 19 |
Hb_011030_020 |
0.2071636359 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 20 |
Hb_004429_070 |
0.2076234476 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |