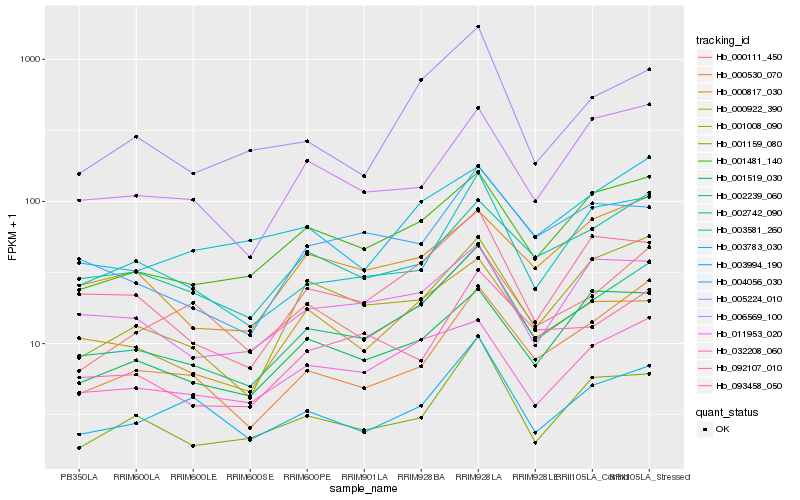
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002239_060 |
0.0 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 2 |
Hb_001481_140 |
0.0917378311 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 3 |
Hb_002742_090 |
0.1058912381 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 4 |
Hb_093458_050 |
0.108975116 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 5 |
Hb_003581_260 |
0.1100637134 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 6 |
Hb_001008_090 |
0.110411447 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 7 |
Hb_000530_070 |
0.1114422698 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 8 |
Hb_032208_060 |
0.1139799074 |
- |
- |
PREDICTED: constitutive photomorphogenesis protein 10 isoform X2 [Populus euphratica] |
| 9 |
Hb_092107_010 |
0.1154625146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005224_010 |
0.1190776891 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 11 |
Hb_000111_450 |
0.1212995852 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 12 |
Hb_000922_390 |
0.1216715149 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 13 |
Hb_003783_030 |
0.1216839089 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 14 |
Hb_003994_190 |
0.1217991616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001159_080 |
0.123082711 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 16 |
Hb_000817_030 |
0.1231223362 |
- |
- |
mak, putative [Ricinus communis] |
| 17 |
Hb_001519_030 |
0.1233574961 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011953_020 |
0.1234132822 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 19 |
Hb_004056_030 |
0.1252039493 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 20 |
Hb_006569_100 |
0.1256550454 |
- |
- |
hypothetical protein POPTR_0005s26990g [Populus trichocarpa] |