| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
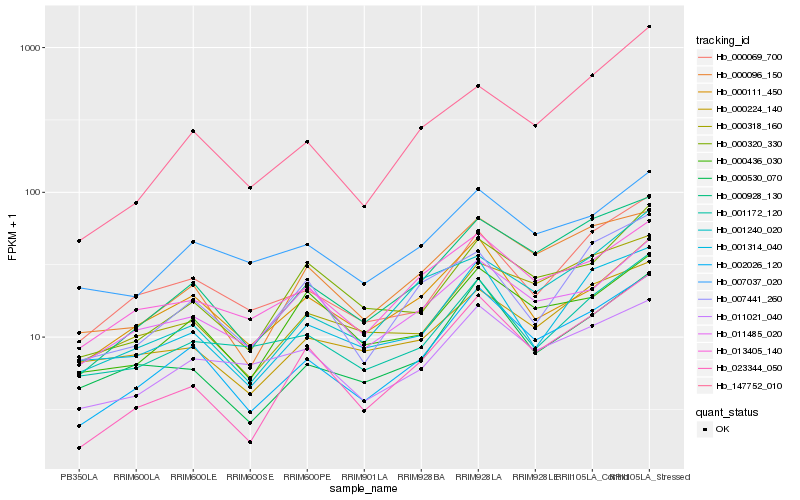
Hb_002026_120 |
0.0 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 2 |
Hb_000928_130 |
0.0634219105 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_000096_150 |
0.0949064154 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 4 |
Hb_023344_050 |
0.0980517582 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 5 |
Hb_000530_070 |
0.0991269304 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 6 |
Hb_000318_160 |
0.1101761961 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |
| 7 |
Hb_001314_040 |
0.1111760665 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 8 |
Hb_013405_140 |
0.1113998309 |
- |
- |
PREDICTED: uncharacterized protein LOC105648437 [Jatropha curcas] |
| 9 |
Hb_007037_020 |
0.1147711149 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 10 |
Hb_001240_020 |
0.1153454612 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 11 |
Hb_000069_700 |
0.1153734475 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 12 |
Hb_147752_010 |
0.1181955964 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 13 |
Hb_001172_120 |
0.1188941363 |
- |
- |
hypothetical protein B456_012G082000 [Gossypium raimondii] |
| 14 |
Hb_000320_330 |
0.1236015238 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |
| 15 |
Hb_000111_450 |
0.1239813325 |
- |
- |
PREDICTED: probable choline kinase 2 [Jatropha curcas] |
| 16 |
Hb_000224_140 |
0.1242888122 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 17 |
Hb_011021_040 |
0.1247169994 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 18 |
Hb_000436_030 |
0.1255508831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007441_260 |
0.1270494463 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_011485_020 |
0.1275042635 |
- |
- |
protein with unknown function [Ricinus communis] |