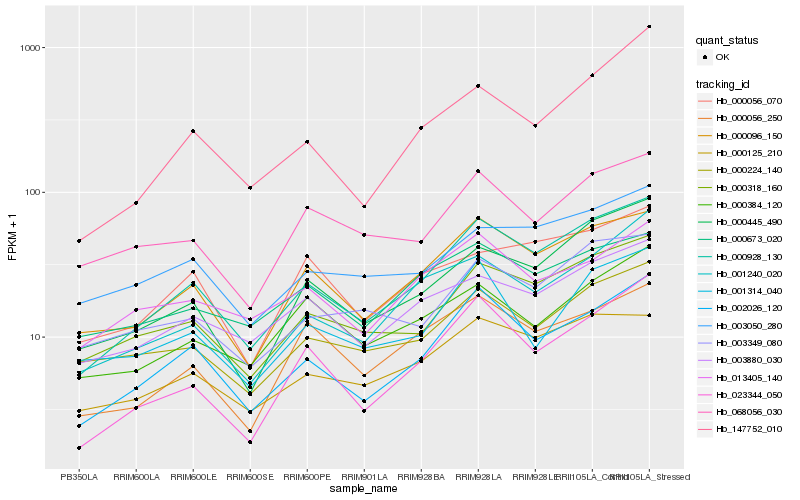
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_130 |
0.0 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 2 |
Hb_002026_120 |
0.0634219105 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 3 |
Hb_000096_150 |
0.0784777681 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 4 |
Hb_000318_160 |
0.0821287866 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |
| 5 |
Hb_000445_490 |
0.0885676192 |
- |
- |
- |
| 6 |
Hb_023344_050 |
0.0974010147 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 7 |
Hb_001240_020 |
0.1001355698 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 8 |
Hb_000125_210 |
0.1033308729 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_147752_010 |
0.1094946787 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 10 |
Hb_003349_080 |
0.1107053867 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 11 |
Hb_003880_030 |
0.1107911937 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000224_140 |
0.1120331203 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 13 |
Hb_000056_250 |
0.1160741776 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_003050_280 |
0.1170115223 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 15 |
Hb_001314_040 |
0.1219990223 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 16 |
Hb_000056_070 |
0.1227035078 |
- |
- |
- |
| 17 |
Hb_068056_030 |
0.124276697 |
- |
- |
ATP synthase epsilon chain, mitochondrial, putative [Ricinus communis] |
| 18 |
Hb_000673_020 |
0.1255083001 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |
| 19 |
Hb_013405_140 |
0.1255336715 |
- |
- |
PREDICTED: uncharacterized protein LOC105648437 [Jatropha curcas] |
| 20 |
Hb_000384_120 |
0.1259739724 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |