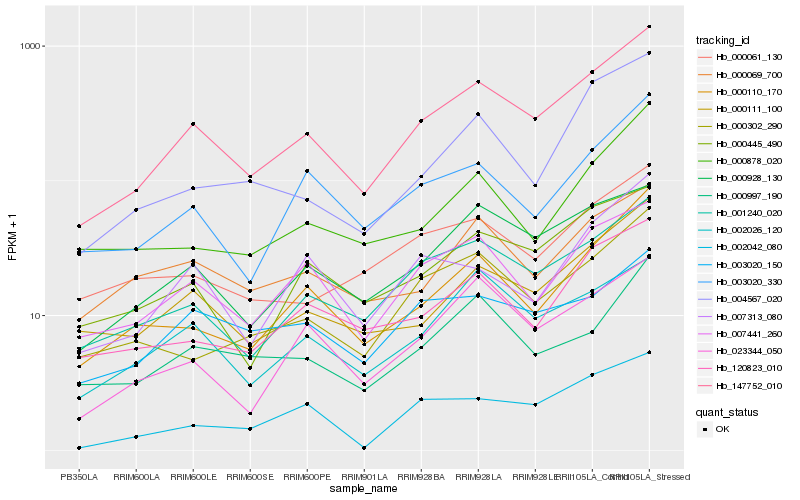
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147752_010 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 2 |
Hb_001240_020 |
0.0929636958 |
- |
- |
PREDICTED: uncharacterized protein LOC105641171 [Jatropha curcas] |
| 3 |
Hb_000111_100 |
0.10356037 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 4 |
Hb_000928_130 |
0.1094946787 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_000110_170 |
0.1123534232 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_007441_260 |
0.1137897599 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_000445_490 |
0.117282333 |
- |
- |
- |
| 8 |
Hb_002026_120 |
0.1181955964 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 9 |
Hb_003020_330 |
0.1187298901 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 10 |
Hb_120823_010 |
0.1214500798 |
- |
- |
- |
| 11 |
Hb_000302_290 |
0.1218600998 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 12 |
Hb_023344_050 |
0.1240817597 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 13 |
Hb_000997_190 |
0.1319981481 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 14 |
Hb_007313_080 |
0.132237055 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 15 |
Hb_000069_700 |
0.132678512 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 16 |
Hb_004567_020 |
0.133217316 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 17 |
Hb_000061_130 |
0.1344902787 |
- |
- |
rotamase, putative [Ricinus communis] |
| 18 |
Hb_002042_080 |
0.1357936833 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 19 |
Hb_000878_020 |
0.1370425164 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 20 |
Hb_003020_150 |
0.1379771572 |
- |
- |
tropinone reductase, putative [Ricinus communis] |