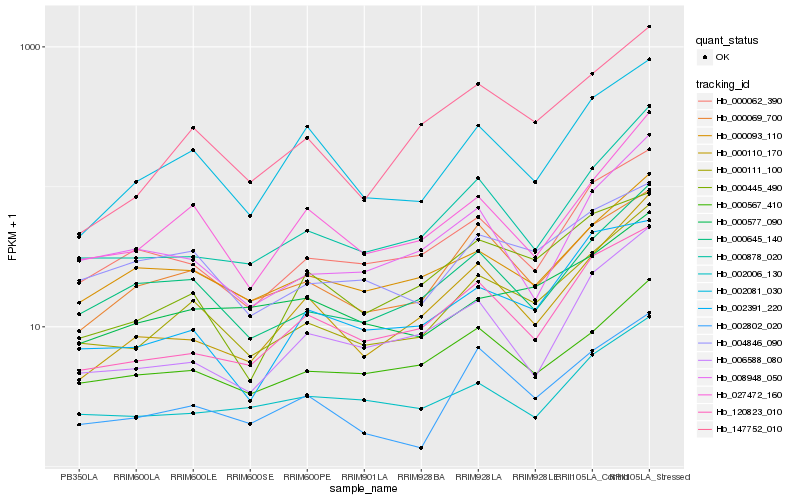
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_100 |
0.0 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 2 |
Hb_000645_140 |
0.0985432894 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A [Nelumbo nucifera] |
| 3 |
Hb_147752_010 |
0.10356037 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 4 |
Hb_000878_020 |
0.1035935132 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 5 |
Hb_027472_160 |
0.1040495102 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 6 |
Hb_000062_390 |
0.1121366363 |
- |
- |
PREDICTED: protein Iojap-related, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000093_110 |
0.1139353202 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 8 |
Hb_120823_010 |
0.1146412458 |
- |
- |
- |
| 9 |
Hb_000069_700 |
0.1164734279 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 10 |
Hb_000567_410 |
0.1204969382 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |
| 11 |
Hb_002081_030 |
0.1212746292 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000110_170 |
0.122003728 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_002802_020 |
0.1248456465 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 14 |
Hb_000445_490 |
0.1249235275 |
- |
- |
- |
| 15 |
Hb_002391_220 |
0.1262698344 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 16 |
Hb_000577_090 |
0.1265819866 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_002006_130 |
0.1279368724 |
- |
- |
PREDICTED: uncharacterized protein LOC105648712 [Jatropha curcas] |
| 18 |
Hb_006588_080 |
0.1289258333 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 19 |
Hb_004846_090 |
0.1289720754 |
transcription factor |
TF Family: HMG |
HMG-box DNA-binding family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_008948_050 |
0.1299379478 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |