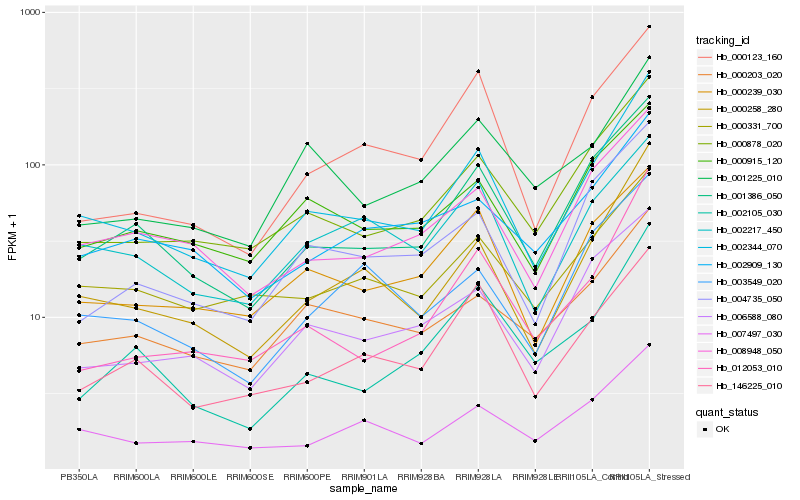
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002344_070 |
0.0 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 2 |
Hb_000258_280 |
0.0690106979 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 3 |
Hb_000878_020 |
0.0892228361 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 4 |
Hb_001386_050 |
0.0916556227 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 5 |
Hb_008948_050 |
0.1063425741 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 6 |
Hb_007497_030 |
0.1076452022 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 7 |
Hb_012053_010 |
0.1170461147 |
- |
- |
srpk, putative [Ricinus communis] |
| 8 |
Hb_000239_030 |
0.117666402 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 9 |
Hb_006588_080 |
0.117970196 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 10 |
Hb_004735_050 |
0.1179721138 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000203_020 |
0.1186340379 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 12 |
Hb_000915_120 |
0.1191821373 |
- |
- |
Peptidylprolyl cis/trans isomerase [Theobroma cacao] |
| 13 |
Hb_146225_010 |
0.1210458345 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 14 |
Hb_000331_700 |
0.1232723307 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 15 |
Hb_000123_160 |
0.1267526638 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 16 |
Hb_002909_130 |
0.1269895944 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_002217_450 |
0.1275000296 |
- |
- |
PREDICTED: probable prefoldin subunit 3 [Jatropha curcas] |
| 18 |
Hb_001225_010 |
0.1289643467 |
- |
- |
unnamed protein product [Coffea canephora] |
| 19 |
Hb_002105_030 |
0.1295111139 |
- |
- |
- |
| 20 |
Hb_003549_020 |
0.1303752684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |