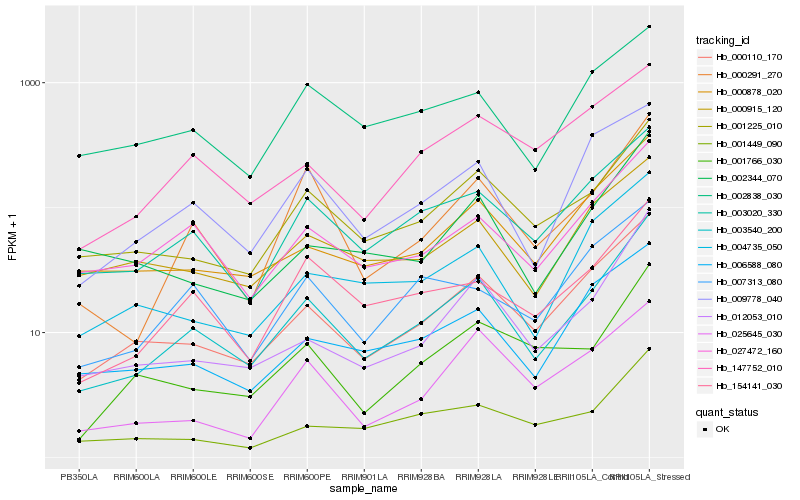
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003540_200 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 2 |
Hb_000110_170 |
0.0958502845 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_003020_330 |
0.1087778405 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 4 |
Hb_001225_010 |
0.1161184611 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000291_270 |
0.1163823769 |
- |
- |
hypothetical protein JCGZ_12006 [Jatropha curcas] |
| 6 |
Hb_012053_010 |
0.1188100469 |
- |
- |
srpk, putative [Ricinus communis] |
| 7 |
Hb_154141_030 |
0.1231155808 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 8 |
Hb_000878_020 |
0.1241813523 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 9 |
Hb_027472_160 |
0.126043134 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 10 |
Hb_009778_040 |
0.1304366121 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 11 |
Hb_006588_080 |
0.1346125589 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 12 |
Hb_004735_050 |
0.1351447761 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001449_090 |
0.1384619261 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007313_080 |
0.1431684065 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 15 |
Hb_001766_030 |
0.1444606632 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 16 |
Hb_002838_030 |
0.144844653 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 17 |
Hb_002344_070 |
0.1468723672 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 18 |
Hb_147752_010 |
0.1485242044 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000915_120 |
0.1510381528 |
- |
- |
Peptidylprolyl cis/trans isomerase [Theobroma cacao] |
| 20 |
Hb_025645_030 |
0.1531626408 |
- |
- |
- |