| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
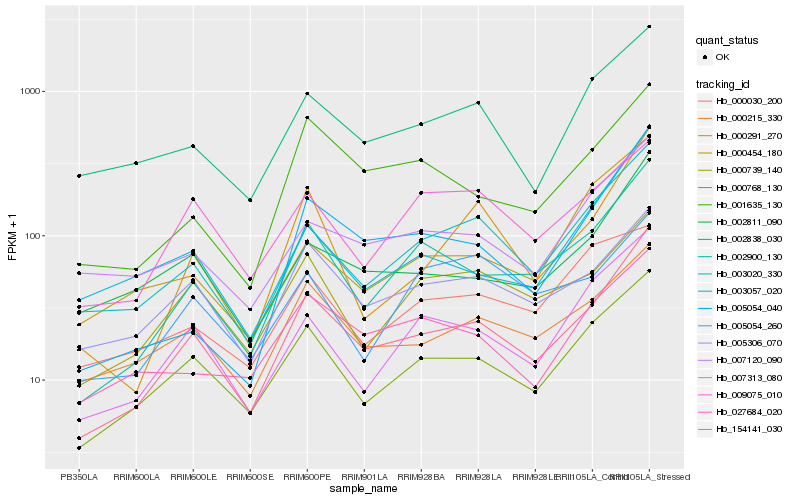
Hb_002900_130 |
0.0 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 2 |
Hb_154141_030 |
0.0904629653 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 3 |
Hb_000768_130 |
0.1103284721 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 4 |
Hb_007313_080 |
0.1109708874 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 5 |
Hb_001635_130 |
0.1269157961 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 6 |
Hb_003020_330 |
0.1284869279 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 7 |
Hb_000739_140 |
0.1369510446 |
- |
- |
PREDICTED: proteasome subunit beta type-1 [Jatropha curcas] |
| 8 |
Hb_005054_040 |
0.1436116173 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 9 |
Hb_005306_070 |
0.1548130455 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 10 |
Hb_003057_020 |
0.1564614372 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 11 |
Hb_000454_180 |
0.1568250014 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 12 |
Hb_027684_020 |
0.1574883659 |
- |
- |
ribosomal protein S10 (mitochondrion) [Nicotiana tabacum] |
| 13 |
Hb_000030_200 |
0.1586591681 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 14 |
Hb_007120_090 |
0.1588985828 |
- |
- |
PREDICTED: 60S ribosomal protein L35 [Jatropha curcas] |
| 15 |
Hb_000215_330 |
0.1603371118 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 16 |
Hb_002811_090 |
0.1608821001 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 17 |
Hb_009075_010 |
0.161678551 |
- |
- |
60S ribosomal protein L10B [Hevea brasiliensis] |
| 18 |
Hb_005054_260 |
0.1628231188 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 19 |
Hb_002838_030 |
0.1636035528 |
- |
- |
40S ribosomal protein S25A [Hevea brasiliensis] |
| 20 |
Hb_000291_270 |
0.1669404346 |
- |
- |
hypothetical protein JCGZ_12006 [Jatropha curcas] |