| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
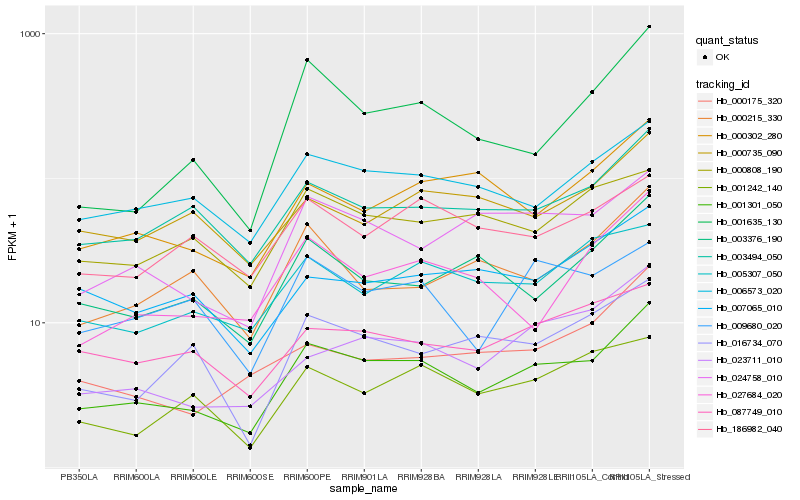
Hb_001301_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_003494_050 |
0.1205987615 |
- |
- |
hypothetical protein PRUPE_ppa013459mg [Prunus persica] |
| 3 |
Hb_006573_020 |
0.1258666189 |
- |
- |
PREDICTED: 40S ribosomal protein S10-like [Jatropha curcas] |
| 4 |
Hb_024758_010 |
0.126579774 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |
| 5 |
Hb_001635_130 |
0.1345337911 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 6 |
Hb_023711_010 |
0.1355713148 |
- |
- |
- |
| 7 |
Hb_000302_280 |
0.1401161385 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 8 |
Hb_003376_190 |
0.1407227896 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 9 |
Hb_027684_020 |
0.1427612774 |
- |
- |
ribosomal protein S10 (mitochondrion) [Nicotiana tabacum] |
| 10 |
Hb_009680_020 |
0.1433964084 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 11 |
Hb_005307_050 |
0.1437024343 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |
| 12 |
Hb_001242_140 |
0.1447439088 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 13 |
Hb_016734_070 |
0.144806898 |
- |
- |
hypothetical_protein [Oryza brachyantha] |
| 14 |
Hb_087749_010 |
0.1448189412 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000808_190 |
0.1464446501 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Eucalyptus grandis] |
| 16 |
Hb_007065_010 |
0.1501075124 |
- |
- |
hypothetical protein Csa_3G379730 [Cucumis sativus] |
| 17 |
Hb_000175_320 |
0.1507712385 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 18 |
Hb_186982_040 |
0.1508256374 |
- |
- |
hypothetical protein PHAVU_003G089200g [Phaseolus vulgaris] |
| 19 |
Hb_000735_090 |
0.1515517757 |
- |
- |
PREDICTED: 40S ribosomal protein S8-like [Gossypium raimondii] |
| 20 |
Hb_000215_330 |
0.1516450667 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |