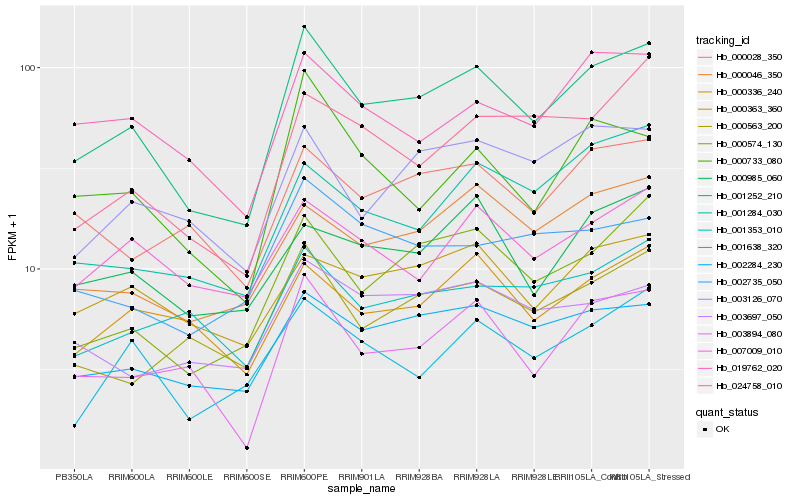
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001252_210 |
0.0 |
- |
- |
PREDICTED: glycine cleavage system H protein 2, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002284_230 |
0.0991509898 |
- |
- |
PREDICTED: uncharacterized protein At5g43822 [Jatropha curcas] |
| 3 |
Hb_000574_130 |
0.102646912 |
- |
- |
calcineurin-like protein [Hevea brasiliensis] |
| 4 |
Hb_003894_080 |
0.1047533121 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Vitis vinifera] |
| 5 |
Hb_000046_350 |
0.1137371154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000733_080 |
0.1156299969 |
- |
- |
PREDICTED: uncharacterized protein LOC105646960 [Jatropha curcas] |
| 7 |
Hb_019762_020 |
0.1169820043 |
- |
- |
PREDICTED: selenoprotein K-like [Nicotiana tomentosiformis] |
| 8 |
Hb_000028_350 |
0.1183710277 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 9 |
Hb_000563_200 |
0.1185598876 |
- |
- |
hypothetical protein JCGZ_18930 [Jatropha curcas] |
| 10 |
Hb_003126_070 |
0.1186633226 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000336_240 |
0.1194051003 |
transcription factor |
TF Family: mTERF |
LOC100285792 [Zea mays] |
| 12 |
Hb_000363_360 |
0.1202827895 |
- |
- |
hypothetical protein PRUPE_ppa009146mg [Prunus persica] |
| 13 |
Hb_001638_320 |
0.123672715 |
- |
- |
hypothetical protein POPTR_0014s10150g [Populus trichocarpa] |
| 14 |
Hb_024758_010 |
0.1240741577 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |
| 15 |
Hb_001353_010 |
0.1241895015 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX11, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000985_060 |
0.1248476002 |
- |
- |
PREDICTED: protein transport protein SFT2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002735_050 |
0.1253878297 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007009_010 |
0.1257004501 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675 [Populus euphratica] |
| 19 |
Hb_003697_050 |
0.125960116 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 20 |
Hb_001284_030 |
0.1260898086 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |