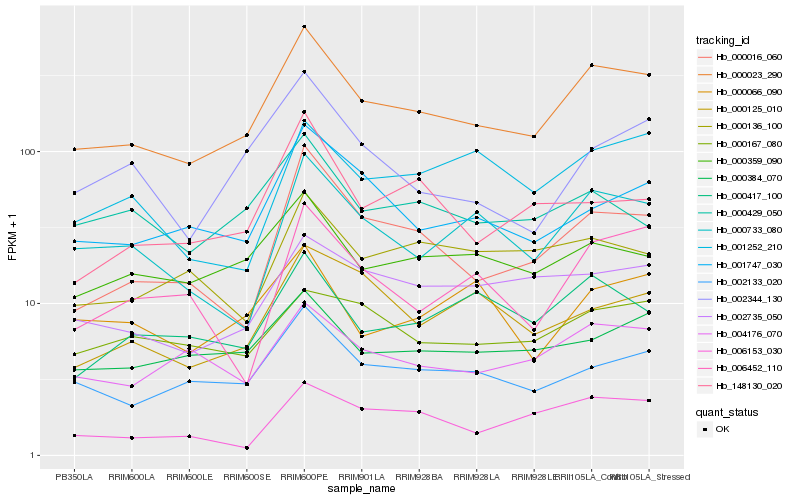
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000023_290 |
0.0 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 2 |
Hb_002133_020 |
0.116184277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000016_060 |
0.1236731434 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_004176_070 |
0.1253652147 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000733_080 |
0.1273412188 |
- |
- |
PREDICTED: uncharacterized protein LOC105646960 [Jatropha curcas] |
| 6 |
Hb_000359_090 |
0.1281283092 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 7 |
Hb_001747_030 |
0.1295371974 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 8 |
Hb_000384_070 |
0.1303350686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002735_050 |
0.1328275238 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000417_100 |
0.1401123482 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 11 |
Hb_002344_130 |
0.1421084044 |
- |
- |
PREDICTED: copper transporter 5 [Jatropha curcas] |
| 12 |
Hb_006153_030 |
0.1440959546 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 13 |
Hb_000167_080 |
0.1473181874 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 14 |
Hb_000429_050 |
0.1474207416 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000136_100 |
0.148631924 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 16 |
Hb_000125_010 |
0.1503075054 |
transcription factor |
TF Family: mTERF |
- |
| 17 |
Hb_000066_090 |
0.1511552707 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 18 |
Hb_006452_110 |
0.1512440879 |
- |
- |
Ubiquinol-cytochrome c reductase complex 7.8 kDa protein, putative [Ricinus communis] |
| 19 |
Hb_148130_020 |
0.1514414206 |
- |
- |
- |
| 20 |
Hb_001252_210 |
0.1525113614 |
- |
- |
PREDICTED: glycine cleavage system H protein 2, mitochondrial [Jatropha curcas] |