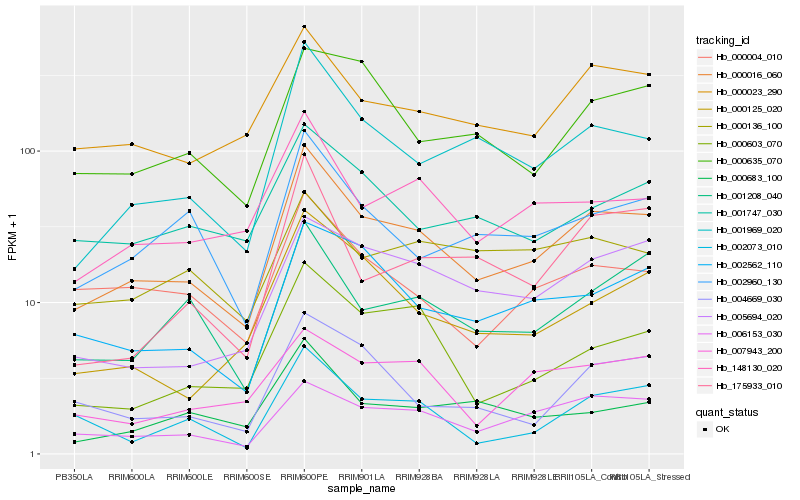
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000016_060 |
0.0 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 2 |
Hb_001969_020 |
0.1198903482 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 3 |
Hb_000023_290 |
0.1236731434 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 4 |
Hb_148130_020 |
0.12856045 |
- |
- |
- |
| 5 |
Hb_006153_030 |
0.1326803301 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 6 |
Hb_002562_110 |
0.1338170467 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002960_130 |
0.1367608685 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 8 |
Hb_001208_040 |
0.1377046926 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 9 |
Hb_000004_010 |
0.137805508 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 10 |
Hb_000125_020 |
0.1425422666 |
transcription factor |
TF Family: mTERF |
- |
| 11 |
Hb_001747_030 |
0.1437611915 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 12 |
Hb_000603_070 |
0.1448084118 |
- |
- |
- |
| 13 |
Hb_004669_030 |
0.1465073272 |
- |
- |
- |
| 14 |
Hb_005694_020 |
0.1489378664 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 15 |
Hb_002073_010 |
0.1509094909 |
- |
- |
- |
| 16 |
Hb_000683_100 |
0.1524044778 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 17 |
Hb_000635_070 |
0.1525556617 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_007943_200 |
0.1566319547 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_175933_010 |
0.1575620831 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 20 |
Hb_000136_100 |
0.1627167701 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |