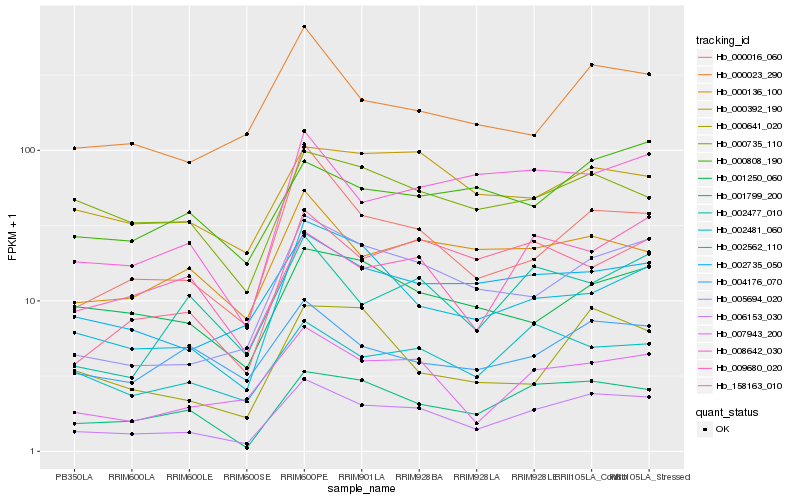
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006153_030 |
0.0 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 2 |
Hb_001799_200 |
0.1156687148 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 3 |
Hb_007943_200 |
0.1211053892 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 4 |
Hb_005694_020 |
0.1285440067 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 5 |
Hb_000016_060 |
0.1326803301 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 6 |
Hb_002735_050 |
0.1327546426 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008642_030 |
0.139242929 |
- |
- |
PREDICTED: UMP-CMP kinase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002562_110 |
0.1428149714 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002481_060 |
0.1437483133 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 10 |
Hb_000023_290 |
0.1440959546 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 11 |
Hb_000136_100 |
0.1449925369 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 12 |
Hb_004176_070 |
0.1488028118 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002477_010 |
0.1492313077 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000735_110 |
0.1494464696 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 15 |
Hb_009680_020 |
0.1502133371 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 16 |
Hb_158163_010 |
0.1506009695 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_000392_190 |
0.1513832732 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 18 |
Hb_001250_060 |
0.1521110469 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 19 |
Hb_000808_190 |
0.1561663838 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Eucalyptus grandis] |
| 20 |
Hb_000641_020 |
0.157213172 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |