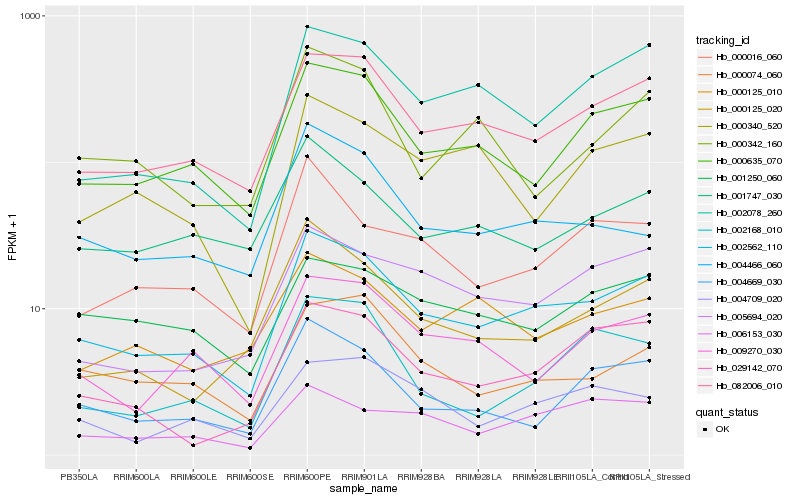
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002562_110 |
0.0 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 2 |
Hb_082006_010 |
0.0897760597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000635_070 |
0.1052335589 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 4 |
Hb_002078_260 |
0.1232334905 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_009270_030 |
0.1304630949 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 6 |
Hb_004669_030 |
0.1327213111 |
- |
- |
- |
| 7 |
Hb_000016_060 |
0.1338170467 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 8 |
Hb_005694_020 |
0.1341441001 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 9 |
Hb_004466_060 |
0.1357016676 |
- |
- |
hypothetical protein POPTR_0009s12110g [Populus trichocarpa] |
| 10 |
Hb_001747_030 |
0.1361644947 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 11 |
Hb_000074_060 |
0.136757341 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 12 |
Hb_000125_020 |
0.1374994041 |
transcription factor |
TF Family: mTERF |
- |
| 13 |
Hb_029142_070 |
0.1401389934 |
- |
- |
- |
| 14 |
Hb_004709_020 |
0.1404132629 |
- |
- |
PREDICTED: putative threonine aspartase [Jatropha curcas] |
| 15 |
Hb_006153_030 |
0.1428149714 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 16 |
Hb_000340_520 |
0.1442854869 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 17 |
Hb_000125_010 |
0.1443376047 |
transcription factor |
TF Family: mTERF |
- |
| 18 |
Hb_001250_060 |
0.1452441517 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 19 |
Hb_000342_160 |
0.148230225 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 20 |
Hb_002168_010 |
0.1511602698 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |