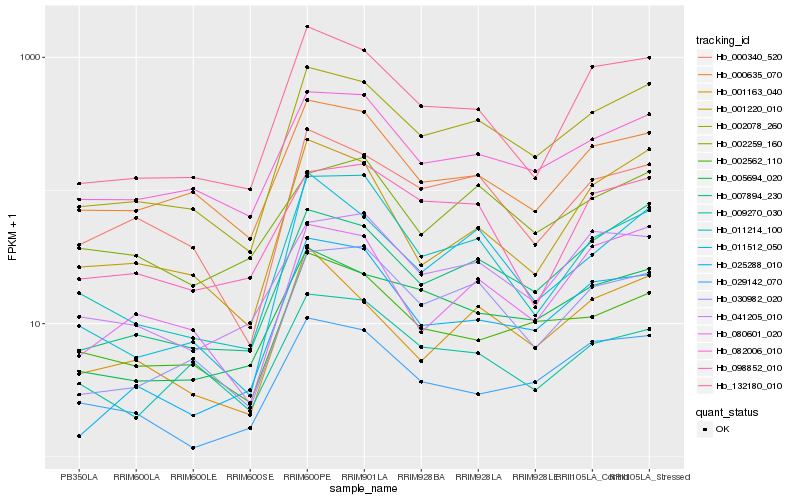
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_260 |
0.0 |
- |
- |
unknown [Populus trichocarpa] |
| 2 |
Hb_007894_230 |
0.0782811819 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 3 |
Hb_030982_020 |
0.0919555006 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_005694_020 |
0.0981225686 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 5 |
Hb_132180_010 |
0.1010137713 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 6 |
Hb_082006_010 |
0.1011179449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000340_520 |
0.1096829253 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 8 |
Hb_041205_010 |
0.1160746384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011214_100 |
0.1178081395 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 10 |
Hb_000635_070 |
0.1190342859 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_011512_050 |
0.1190487904 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 12 |
Hb_080601_020 |
0.119346596 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 13 |
Hb_002562_110 |
0.1232334905 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 14 |
Hb_029142_070 |
0.1258089729 |
- |
- |
- |
| 15 |
Hb_001163_040 |
0.1271547507 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 16 |
Hb_001220_010 |
0.1301825622 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 17 |
Hb_025288_010 |
0.1304133962 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 18 |
Hb_098852_010 |
0.1316923171 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 19 |
Hb_002259_160 |
0.1393660968 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |
| 20 |
Hb_009270_030 |
0.13961364 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |