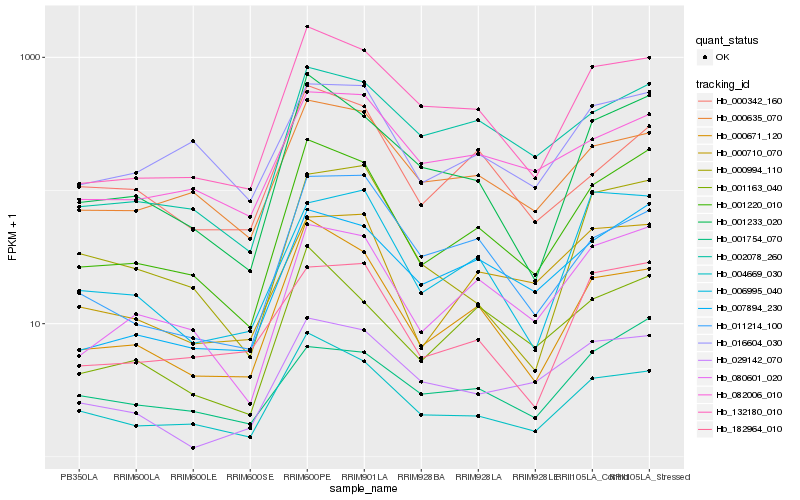
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001220_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 2 |
Hb_080601_020 |
0.0867003529 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 3 |
Hb_001233_020 |
0.1018094824 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 4 |
Hb_132180_010 |
0.1157663817 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 5 |
Hb_000671_120 |
0.1161939518 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 6 |
Hb_004669_030 |
0.1168764332 |
- |
- |
- |
| 7 |
Hb_007894_230 |
0.1186930081 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 8 |
Hb_001163_040 |
0.1298889061 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 9 |
Hb_002078_260 |
0.1301825622 |
- |
- |
unknown [Populus trichocarpa] |
| 10 |
Hb_000635_070 |
0.1358528945 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_182964_010 |
0.137987774 |
- |
- |
PREDICTED: uncharacterized protein LOC105642044 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000710_070 |
0.1383074016 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 13 |
Hb_016604_030 |
0.1430171558 |
- |
- |
histone H4 [Zea mays] |
| 14 |
Hb_001754_070 |
0.1489646318 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 15 |
Hb_029142_070 |
0.1511785509 |
- |
- |
- |
| 16 |
Hb_006995_040 |
0.1514786219 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 17 |
Hb_082006_010 |
0.1525614951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011214_100 |
0.1544552404 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 19 |
Hb_000342_160 |
0.1550006756 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 20 |
Hb_000994_110 |
0.1553079887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |