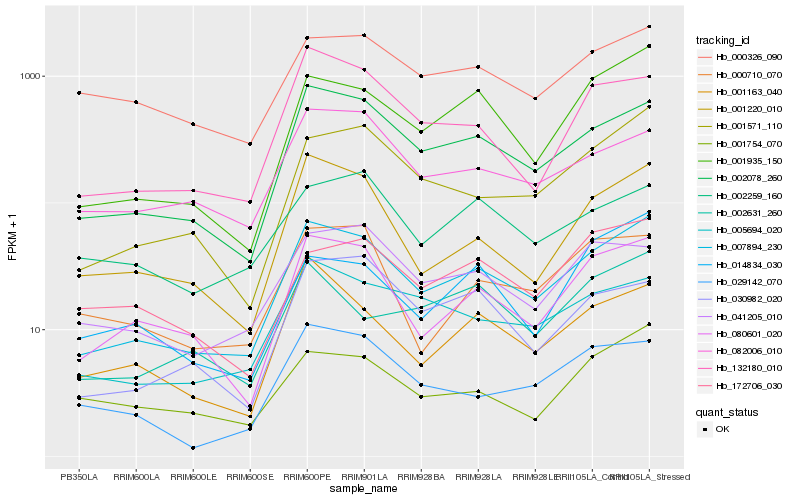
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_230 |
0.0 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 2 |
Hb_002078_260 |
0.0782811819 |
- |
- |
unknown [Populus trichocarpa] |
| 3 |
Hb_080601_020 |
0.1060320315 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 4 |
Hb_001935_150 |
0.1073307912 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 5 |
Hb_001571_110 |
0.1125022478 |
- |
- |
PREDICTED: 60S ribosomal protein L44-like [Gossypium raimondii] |
| 6 |
Hb_001220_010 |
0.1186930081 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 7 |
Hb_005694_020 |
0.121953424 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 8 |
Hb_132180_010 |
0.1247247094 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 9 |
Hb_001163_040 |
0.1307901735 |
- |
- |
Cytochrome-c oxidases,electron carriers [Theobroma cacao] |
| 10 |
Hb_029142_070 |
0.1350761763 |
- |
- |
- |
| 11 |
Hb_041205_010 |
0.1368211703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_082006_010 |
0.1391367066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_030982_020 |
0.1421616568 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_001754_070 |
0.1438345079 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 15 |
Hb_000710_070 |
0.144787562 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 16 |
Hb_000326_090 |
0.1455850691 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 17 |
Hb_172706_030 |
0.1465510837 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 18 |
Hb_002631_260 |
0.1491821308 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 19 |
Hb_002259_160 |
0.1514368479 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |
| 20 |
Hb_014834_030 |
0.1515564245 |
- |
- |
- |