| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
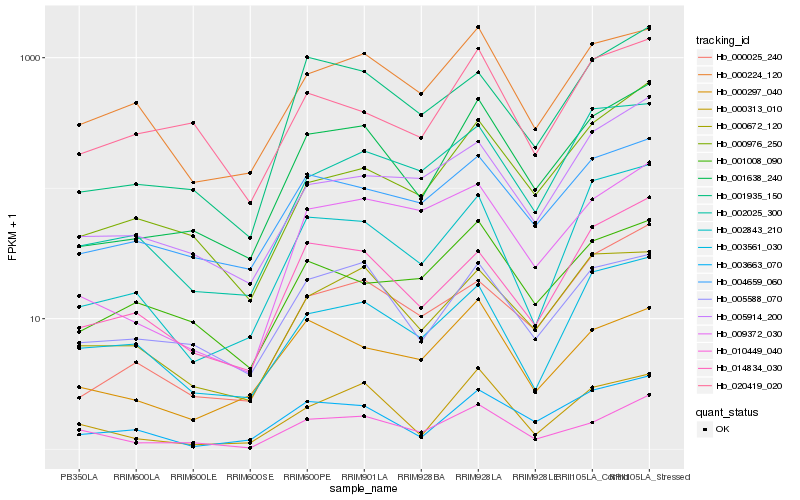
Hb_001638_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 2 |
Hb_003663_070 |
0.1157881176 |
- |
- |
- |
| 3 |
Hb_001935_150 |
0.1198280143 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 4 |
Hb_004659_060 |
0.1245463672 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 5 |
Hb_002843_210 |
0.1249017274 |
- |
- |
PREDICTED: uncharacterized protein LOC105647178 [Jatropha curcas] |
| 6 |
Hb_005588_070 |
0.1277562541 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 7 |
Hb_000672_120 |
0.1282677183 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 8 |
Hb_014834_030 |
0.1305592226 |
- |
- |
- |
| 9 |
Hb_000025_240 |
0.1344844494 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 10 |
Hb_000313_010 |
0.1356890944 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 11 |
Hb_000976_250 |
0.1364577845 |
- |
- |
- |
| 12 |
Hb_000224_120 |
0.1383648429 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 13 |
Hb_009372_030 |
0.1398247912 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 14 |
Hb_002025_300 |
0.1400284105 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 15 |
Hb_010449_040 |
0.145282229 |
- |
- |
PREDICTED: probable CDP-diacylglycerol--inositol 3-phosphatidyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_005914_200 |
0.1472464978 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 17 |
Hb_000297_040 |
0.1483663013 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 18 |
Hb_001008_090 |
0.1492392164 |
- |
- |
cytochrome C oxidase, putative [Ricinus communis] |
| 19 |
Hb_020419_020 |
0.1502329014 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 20 |
Hb_003561_030 |
0.1512828782 |
- |
- |
PREDICTED: uncharacterized protein LOC105645274 [Jatropha curcas] |