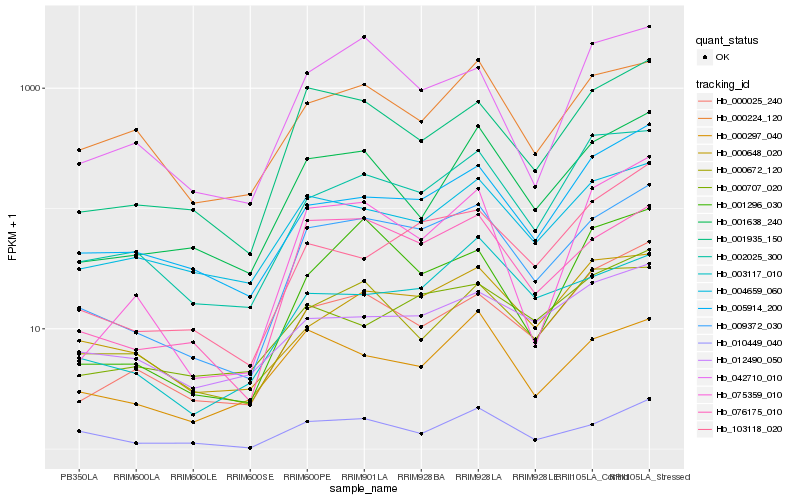
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009372_030 |
0.0 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 2 |
Hb_076175_010 |
0.0867576331 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB7 [Cucumis melo] |
| 3 |
Hb_001935_150 |
0.122876246 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Jatropha curcas] |
| 4 |
Hb_002025_300 |
0.1333957266 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 5 |
Hb_010449_040 |
0.1383289726 |
- |
- |
PREDICTED: probable CDP-diacylglycerol--inositol 3-phosphatidyltransferase 2 [Jatropha curcas] |
| 6 |
Hb_001638_240 |
0.1398247912 |
- |
- |
PREDICTED: uncharacterized protein LOC100262493 [Vitis vinifera] |
| 7 |
Hb_004659_060 |
0.1409657226 |
- |
- |
RAS-related GTP-binding family protein [Populus trichocarpa] |
| 8 |
Hb_000648_020 |
0.1429972069 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 9 |
Hb_000025_240 |
0.1449593339 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 10 |
Hb_005914_200 |
0.1464442311 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 11 |
Hb_075359_010 |
0.1495340903 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 12 |
Hb_000224_120 |
0.1502576518 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 13 |
Hb_003117_010 |
0.1510728302 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 14 |
Hb_042710_010 |
0.1523472031 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 15 |
Hb_000672_120 |
0.1528414309 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 16 |
Hb_103118_020 |
0.1541819271 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 17 |
Hb_000707_020 |
0.1553695389 |
- |
- |
hypothetical protein B456_002G255500 [Gossypium raimondii] |
| 18 |
Hb_001296_030 |
0.1565872333 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |
| 19 |
Hb_000297_040 |
0.1567468759 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 20 |
Hb_012490_050 |
0.1570366563 |
- |
- |
PREDICTED: uncharacterized protein LOC105637921 [Jatropha curcas] |