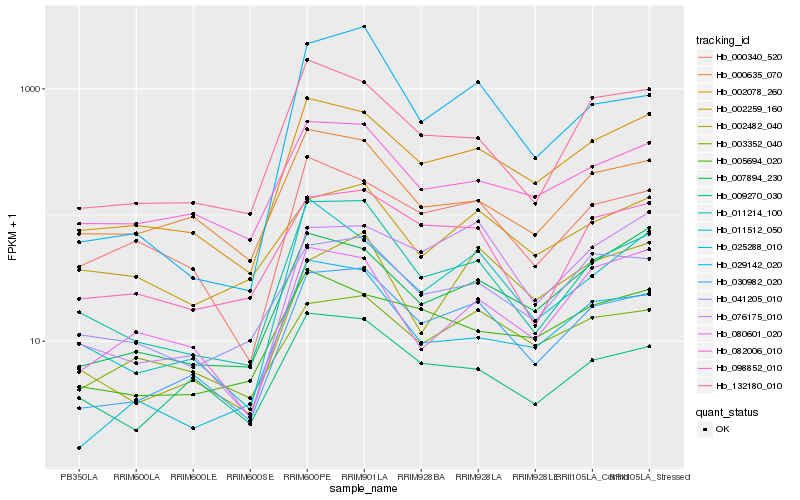
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030982_020 |
0.0 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 2 |
Hb_002078_260 |
0.0919555006 |
- |
- |
unknown [Populus trichocarpa] |
| 3 |
Hb_011214_100 |
0.1142670177 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 4 |
Hb_098852_010 |
0.1220831039 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 5 |
Hb_041205_010 |
0.1284616302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_132180_010 |
0.1291776532 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 7 |
Hb_009270_030 |
0.134075251 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 8 |
Hb_082006_010 |
0.1353758538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000340_520 |
0.1378372164 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 10 |
Hb_076175_010 |
0.1391206782 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB7 [Cucumis melo] |
| 11 |
Hb_007894_230 |
0.1421616568 |
- |
- |
30S ribosomal protein S14, putative [Ricinus communis] |
| 12 |
Hb_025288_010 |
0.1458637471 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 13 |
Hb_000635_070 |
0.1462884454 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_003352_040 |
0.1502521416 |
- |
- |
PREDICTED: 26S proteasome regulatory subunit RPN13 [Jatropha curcas] |
| 15 |
Hb_005694_020 |
0.15037944 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 16 |
Hb_011512_050 |
0.1539552111 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 13-A isoform 2 [Theobroma cacao] |
| 17 |
Hb_002259_160 |
0.1560560287 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |
| 18 |
Hb_029142_020 |
0.1583480481 |
- |
- |
hypothetical protein POPTR_0004s23900g [Populus trichocarpa] |
| 19 |
Hb_002482_040 |
0.1616851474 |
- |
- |
PREDICTED: uncharacterized protein LOC105642409 [Jatropha curcas] |
| 20 |
Hb_080601_020 |
0.1622111976 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |