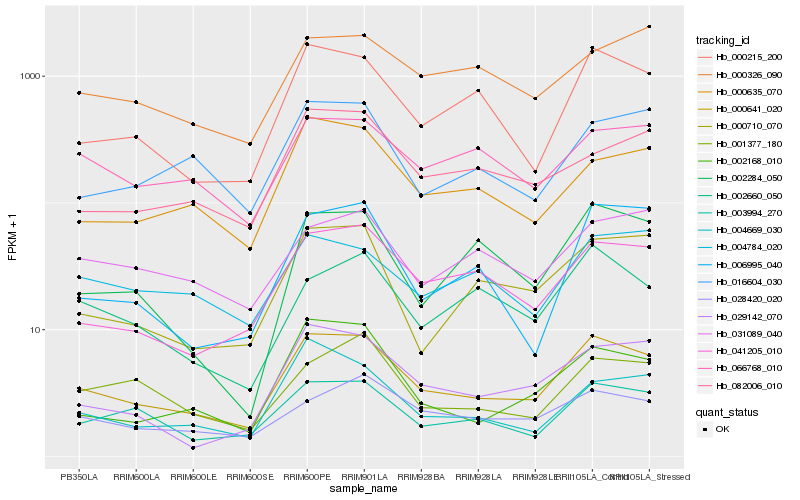
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000641_020 |
0.0 |
- |
- |
PREDICTED: tubulin-folding cofactor B-like [Jatropha curcas] |
| 2 |
Hb_000215_200 |
0.1142335284 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 3 |
Hb_003994_270 |
0.1142472792 |
- |
- |
PREDICTED: uncharacterized protein LOC105646152 [Jatropha curcas] |
| 4 |
Hb_041205_010 |
0.1194730901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000635_070 |
0.1280222268 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 6 |
Hb_029142_070 |
0.1294493187 |
- |
- |
- |
| 7 |
Hb_000710_070 |
0.1330592429 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 8 |
Hb_028420_020 |
0.1355612433 |
- |
- |
PREDICTED: uncharacterized protein LOC105647566 [Jatropha curcas] |
| 9 |
Hb_002168_010 |
0.1377895791 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 10 |
Hb_066768_010 |
0.1382223705 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_004669_030 |
0.13996102 |
- |
- |
- |
| 12 |
Hb_016604_030 |
0.1403842744 |
- |
- |
histone H4 [Zea mays] |
| 13 |
Hb_082006_010 |
0.1426440328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002660_050 |
0.1429009774 |
- |
- |
PREDICTED: uncharacterized protein LOC105642218 [Jatropha curcas] |
| 15 |
Hb_001377_180 |
0.1433933302 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1 [Jatropha curcas] |
| 16 |
Hb_006995_040 |
0.1453475094 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 17 |
Hb_004784_020 |
0.1466617456 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 18 |
Hb_000326_090 |
0.1475632303 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 19 |
Hb_031089_040 |
0.1480187444 |
- |
- |
thioredoxin H-type 1 [Hevea brasiliensis] |
| 20 |
Hb_002284_050 |
0.1486226165 |
- |
- |
hypothetical protein POPTR_0002s00740g [Populus trichocarpa] |