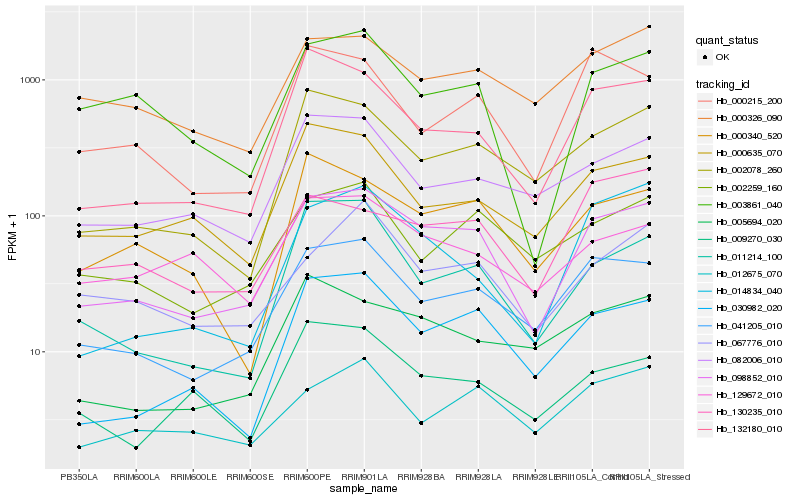
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098852_010 |
0.0 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 2 |
Hb_041205_010 |
0.099781318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_030982_020 |
0.1220831039 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_132180_010 |
0.1291701638 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 5 |
Hb_002078_260 |
0.1316923171 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_003861_040 |
0.1368170597 |
- |
- |
hypothetical protein JCGZ_16325 [Jatropha curcas] |
| 7 |
Hb_000326_090 |
0.1369230974 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 8 |
Hb_002259_160 |
0.1375868674 |
- |
- |
hypothetical protein PSEUBRA_SCAF14g01508 [Pseudozyma brasiliensis GHG001] |
| 9 |
Hb_082006_010 |
0.1388670965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011214_100 |
0.1407285428 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 11 |
Hb_000340_520 |
0.1417663048 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 12 |
Hb_000635_070 |
0.1419619101 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 13 |
Hb_005694_020 |
0.1429134943 |
- |
- |
PREDICTED: ORM1-like protein 2 [Gossypium raimondii] |
| 14 |
Hb_012675_070 |
0.1433461715 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Pyrus x bretschneideri] |
| 15 |
Hb_067776_010 |
0.1454380518 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 16 |
Hb_009270_030 |
0.147477812 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 17 |
Hb_000215_200 |
0.1476380799 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 18 |
Hb_014834_040 |
0.1494864963 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 19 |
Hb_129672_010 |
0.1519607987 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 20 |
Hb_130235_010 |
0.1522907475 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |