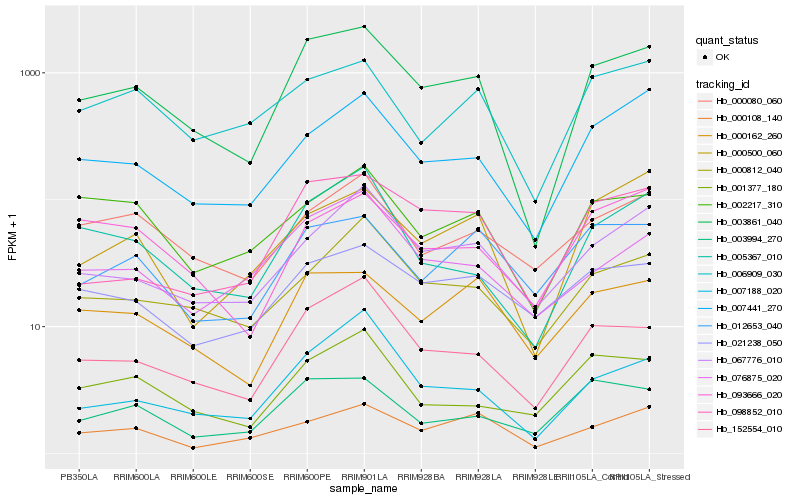
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003861_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_16325 [Jatropha curcas] |
| 2 |
Hb_152554_010 |
0.1110085846 |
- |
- |
hypothetical protein B456_008G095800, partial [Gossypium raimondii] |
| 3 |
Hb_000162_260 |
0.1342932182 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-1 [Nelumbo nucifera] |
| 4 |
Hb_005367_010 |
0.1344636595 |
- |
- |
- |
| 5 |
Hb_007441_270 |
0.1360232104 |
- |
- |
PREDICTED: 40S ribosomal protein S11 [Jatropha curcas] |
| 6 |
Hb_098852_010 |
0.1368170597 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 7 |
Hb_000812_040 |
0.1438919285 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 8 |
Hb_067776_010 |
0.1444454662 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 9 |
Hb_003994_270 |
0.1451638359 |
- |
- |
PREDICTED: uncharacterized protein LOC105646152 [Jatropha curcas] |
| 10 |
Hb_006909_030 |
0.1463891755 |
- |
- |
40S ribosomal protein S17B [Hevea brasiliensis] |
| 11 |
Hb_007188_020 |
0.1485029966 |
- |
- |
hypothetical protein PRUPE_ppb010436mg [Prunus persica] |
| 12 |
Hb_000500_060 |
0.1510044505 |
- |
- |
BnaC08g21190D [Brassica napus] |
| 13 |
Hb_076875_020 |
0.1525601653 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 14 |
Hb_000108_140 |
0.1527205395 |
- |
- |
hypothetical protein CICLE_v10019690mg [Citrus clementina] |
| 15 |
Hb_002217_310 |
0.1531638384 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 16 |
Hb_012653_040 |
0.1550465445 |
- |
- |
UV-induced protein uvi31, putative [Ricinus communis] |
| 17 |
Hb_093666_020 |
0.1550682086 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 18 |
Hb_021238_050 |
0.1560152668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001377_180 |
0.1567274196 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1 [Jatropha curcas] |
| 20 |
Hb_000080_060 |
0.1569437143 |
- |
- |
RNA polymerase I, II and III subunit RPB8, partial [Manihot esculenta] |