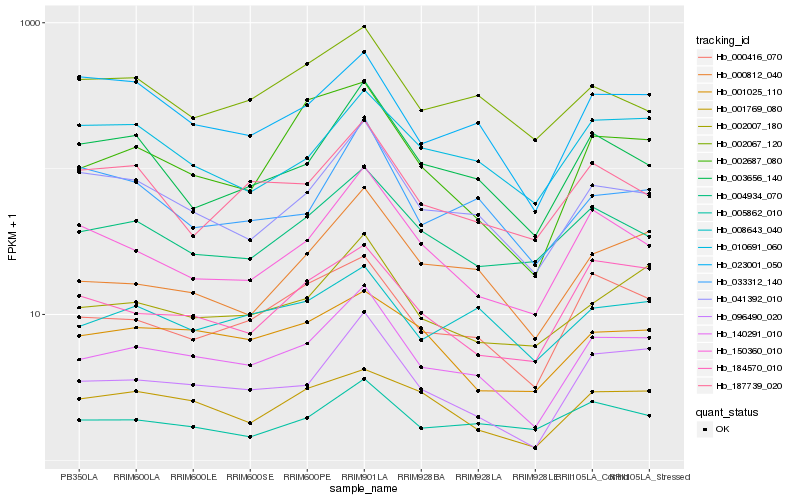
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140291_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17222 [Jatropha curcas] |
| 2 |
Hb_096490_020 |
0.0891044965 |
- |
- |
PREDICTED: UPF0565 protein C2orf69 [Jatropha curcas] |
| 3 |
Hb_041392_010 |
0.0995212188 |
- |
- |
PREDICTED: DNA-binding protein BIN4 [Jatropha curcas] |
| 4 |
Hb_002007_180 |
0.1029058819 |
- |
- |
PREDICTED: uncharacterized protein LOC105647525 [Jatropha curcas] |
| 5 |
Hb_000416_070 |
0.1066494741 |
- |
- |
hypothetical protein JCGZ_21620 [Jatropha curcas] |
| 6 |
Hb_000812_040 |
0.1157870026 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 7 |
Hb_003656_140 |
0.1192913566 |
- |
- |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_150360_010 |
0.1222762631 |
- |
- |
hypothetical protein JCGZ_16890 [Jatropha curcas] |
| 9 |
Hb_023001_050 |
0.1234933362 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 10 |
Hb_004934_070 |
0.1236792428 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 11 |
Hb_002687_080 |
0.1261846067 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 12 |
Hb_008643_040 |
0.12625284 |
- |
- |
PREDICTED: jacalin-related lectin 3-like [Jatropha curcas] |
| 13 |
Hb_001025_110 |
0.1267485862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_184570_010 |
0.127102652 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 15 |
Hb_001769_080 |
0.1273936143 |
- |
- |
hypothetical protein SORBIDRAFT_10g023200 [Sorghum bicolor] |
| 16 |
Hb_005862_010 |
0.1317528397 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |
| 17 |
Hb_033312_140 |
0.13372154 |
- |
- |
RNA and export factor-binding family protein [Populus trichocarpa] |
| 18 |
Hb_187739_020 |
0.1339713232 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 19 |
Hb_010691_060 |
0.1356941453 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 20 |
Hb_002067_120 |
0.1358517464 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |