| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
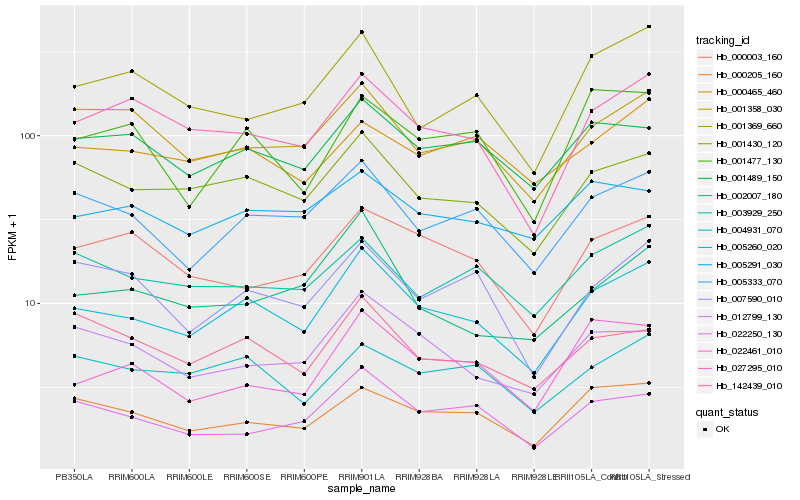
Hb_005260_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633789 [Jatropha curcas] |
| 2 |
Hb_001430_120 |
0.0831309879 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 3 |
Hb_005333_070 |
0.0886050335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_027295_010 |
0.0975597806 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 5 |
Hb_000205_160 |
0.1041357773 |
- |
- |
Peroxisome biogenesis protein 12 [Morus notabilis] |
| 6 |
Hb_001489_150 |
0.1064401428 |
- |
- |
PREDICTED: uncharacterized protein LOC105123623 isoform X2 [Populus euphratica] |
| 7 |
Hb_001358_030 |
0.1073184177 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 8 |
Hb_012799_130 |
0.1092075871 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 9 |
Hb_004931_070 |
0.1104777999 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007590_010 |
0.1122806521 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_003929_250 |
0.113977762 |
- |
- |
PREDICTED: proteasome assembly chaperone 4 [Jatropha curcas] |
| 12 |
Hb_142439_010 |
0.1140812192 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 13 |
Hb_002007_180 |
0.1143794906 |
- |
- |
PREDICTED: uncharacterized protein LOC105647525 [Jatropha curcas] |
| 14 |
Hb_022250_130 |
0.1143841619 |
- |
- |
inositol hexaphosphate kinase, putative [Ricinus communis] |
| 15 |
Hb_000003_160 |
0.1147163508 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 16 |
Hb_001369_660 |
0.1159524946 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 17 |
Hb_001477_130 |
0.1169131868 |
- |
- |
PREDICTED: uncharacterized protein LOC105646871 [Jatropha curcas] |
| 18 |
Hb_022461_010 |
0.1177442343 |
- |
- |
- |
| 19 |
Hb_000465_460 |
0.1181130798 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 [Jatropha curcas] |
| 20 |
Hb_005291_030 |
0.1185695392 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |