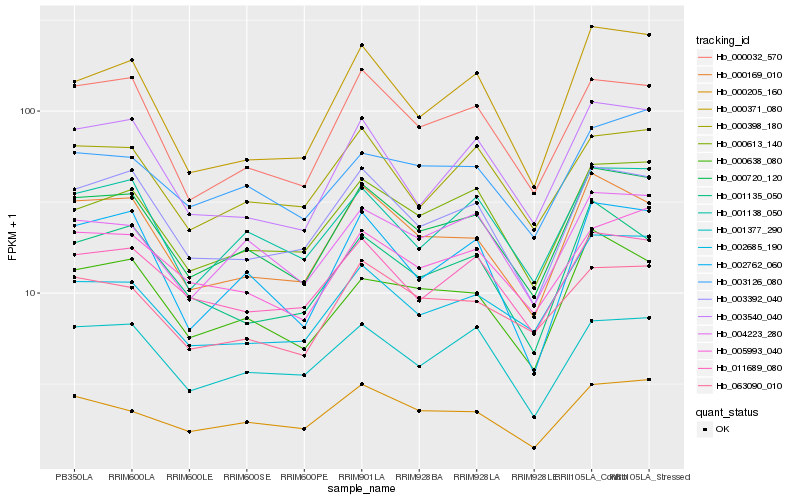
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000720_120 |
0.0 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |
| 2 |
Hb_000169_010 |
0.0553780569 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_001138_050 |
0.0602442195 |
- |
- |
hypothetical protein JCGZ_15696 [Jatropha curcas] |
| 4 |
Hb_003392_040 |
0.0604295003 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Jatropha curcas] |
| 5 |
Hb_000613_140 |
0.061864156 |
- |
- |
PREDICTED: indole-3-glycerol phosphate synthase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000638_080 |
0.0648593959 |
- |
- |
PREDICTED: uncharacterized protein LOC105632765 [Jatropha curcas] |
| 7 |
Hb_000032_570 |
0.064996105 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: RNA polymerase II transcriptional coactivator KELP [Jatropha curcas] |
| 8 |
Hb_004223_280 |
0.0672797602 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 6 [Jatropha curcas] |
| 9 |
Hb_002762_060 |
0.0706922708 |
- |
- |
PREDICTED: diphthine methyltransferase homolog [Jatropha curcas] |
| 10 |
Hb_011689_080 |
0.0710655112 |
- |
- |
PREDICTED: uncharacterized protein LOC105631867 [Jatropha curcas] |
| 11 |
Hb_003540_040 |
0.0716448454 |
- |
- |
hypothetical protein JCGZ_20979 [Jatropha curcas] |
| 12 |
Hb_005993_040 |
0.0717106168 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit wdr4 [Jatropha curcas] |
| 13 |
Hb_001377_290 |
0.0731623914 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase accessory protein, mitochondrial [Jatropha curcas] |
| 14 |
Hb_063090_010 |
0.073693902 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000205_160 |
0.0738078014 |
- |
- |
Peroxisome biogenesis protein 12 [Morus notabilis] |
| 16 |
Hb_000371_080 |
0.0751317183 |
- |
- |
ubiquitin-conjugating enzyme m, putative [Ricinus communis] |
| 17 |
Hb_000398_180 |
0.0757162499 |
- |
- |
hypothetical protein PRUPE_ppa009665mg [Prunus persica] |
| 18 |
Hb_001135_050 |
0.0783115324 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002685_190 |
0.0795604243 |
transcription factor |
TF Family: SET |
PREDICTED: probable zinc metalloprotease EGY1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003126_080 |
0.0795844358 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |