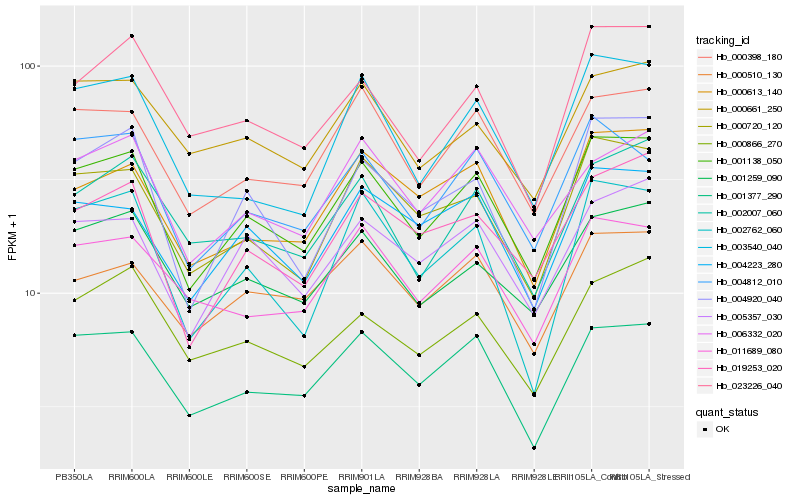
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001138_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_15696 [Jatropha curcas] |
| 2 |
Hb_000398_180 |
0.059446671 |
- |
- |
hypothetical protein PRUPE_ppa009665mg [Prunus persica] |
| 3 |
Hb_000720_120 |
0.0602442195 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |
| 4 |
Hb_001377_290 |
0.0606034628 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase accessory protein, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005357_030 |
0.0642401691 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase regulatory protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001259_090 |
0.0664998965 |
- |
- |
hypothetical protein B456_005G056700 [Gossypium raimondii] |
| 7 |
Hb_019253_020 |
0.0668403074 |
- |
- |
PREDICTED: uncharacterized protein LOC105647727 [Jatropha curcas] |
| 8 |
Hb_004812_010 |
0.068605703 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD6 [Jatropha curcas] |
| 9 |
Hb_004920_040 |
0.0699727979 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 10 |
Hb_004223_280 |
0.0711447536 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 6 [Jatropha curcas] |
| 11 |
Hb_000613_140 |
0.071723765 |
- |
- |
PREDICTED: indole-3-glycerol phosphate synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006332_020 |
0.0733945217 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 13 |
Hb_003540_040 |
0.0738995739 |
- |
- |
hypothetical protein JCGZ_20979 [Jatropha curcas] |
| 14 |
Hb_002007_060 |
0.0753483129 |
- |
- |
hypothetical protein JCGZ_23323 [Jatropha curcas] |
| 15 |
Hb_000866_270 |
0.0753948978 |
- |
- |
PREDICTED: uncharacterized protein LOC105642993 [Jatropha curcas] |
| 16 |
Hb_002762_060 |
0.0765251999 |
- |
- |
PREDICTED: diphthine methyltransferase homolog [Jatropha curcas] |
| 17 |
Hb_000661_250 |
0.076647614 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 18 |
Hb_011689_080 |
0.077903585 |
- |
- |
PREDICTED: uncharacterized protein LOC105631867 [Jatropha curcas] |
| 19 |
Hb_023226_040 |
0.0781894407 |
- |
- |
PREDICTED: kxDL motif-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_000510_130 |
0.078615467 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |