| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
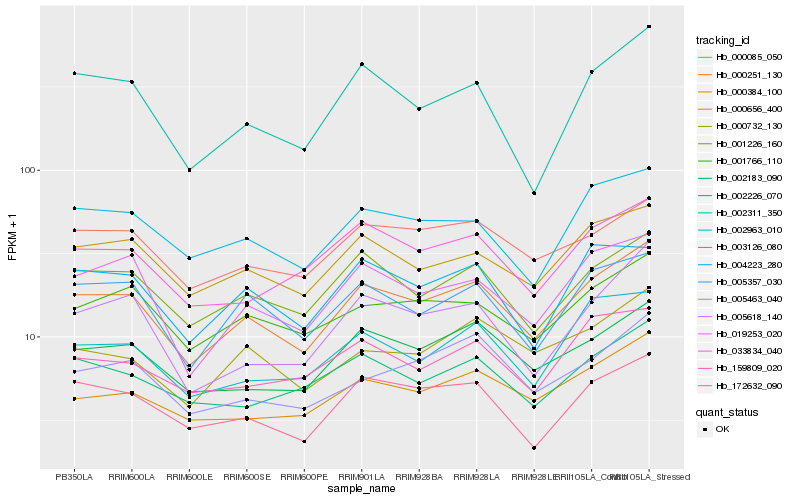
Hb_000251_130 |
0.0 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 2 |
Hb_001226_160 |
0.0671682974 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein B''-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000656_400 |
0.0701103649 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 4 |
Hb_002183_090 |
0.0702842094 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 5 |
Hb_005357_030 |
0.070977079 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase regulatory protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_172632_090 |
0.0716674515 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 7 |
Hb_019253_020 |
0.0737915419 |
- |
- |
PREDICTED: uncharacterized protein LOC105647727 [Jatropha curcas] |
| 8 |
Hb_003126_080 |
0.0743558877 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 9 |
Hb_001766_110 |
0.0751245483 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 10 |
Hb_000384_100 |
0.0775847589 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 11 |
Hb_000085_050 |
0.0793743126 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_002311_350 |
0.080791051 |
- |
- |
PREDICTED: 60S ribosomal protein L19-3 [Jatropha curcas] |
| 13 |
Hb_033834_040 |
0.080812765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002226_070 |
0.0833604682 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002963_010 |
0.0838596179 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000732_130 |
0.0839266071 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_17108 [Jatropha curcas] |
| 17 |
Hb_005618_140 |
0.0849170375 |
- |
- |
PREDICTED: uncharacterized protein LOC105642706 [Jatropha curcas] |
| 18 |
Hb_004223_280 |
0.0853911123 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 6 [Jatropha curcas] |
| 19 |
Hb_005463_040 |
0.0869728976 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 20 |
Hb_159809_020 |
0.087150388 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |