| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
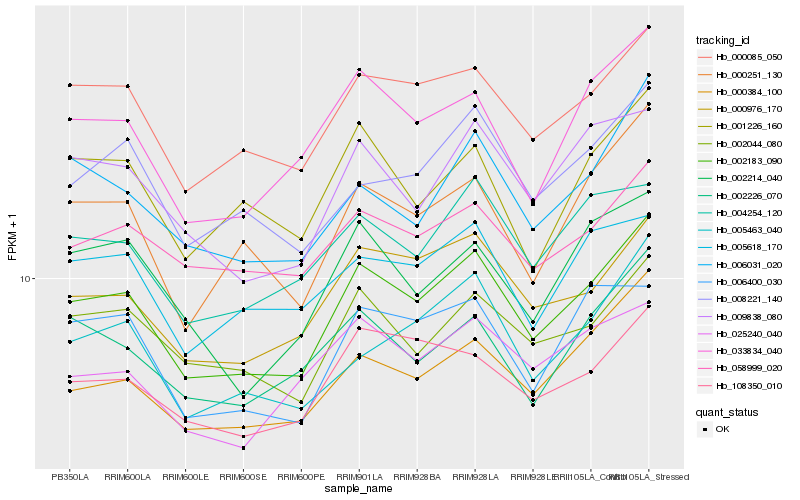
Hb_002183_090 |
0.0 |
- |
- |
PREDICTED: protein EMSY-LIKE 1 [Jatropha curcas] |
| 2 |
Hb_000085_050 |
0.0566703355 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_000976_170 |
0.0605262053 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 4 |
Hb_000384_100 |
0.0650518507 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 5 |
Hb_033834_040 |
0.0659576763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002044_080 |
0.068071737 |
- |
- |
PREDICTED: secretion-regulating guanine nucleotide exchange factor isoform X2 [Jatropha curcas] |
| 7 |
Hb_000251_130 |
0.0702842094 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 8 |
Hb_009838_080 |
0.0711028684 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 9 |
Hb_004254_120 |
0.0731723234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_108350_010 |
0.0737288284 |
- |
- |
GTP-binding protein erg, putative [Ricinus communis] |
| 11 |
Hb_002226_070 |
0.0745513276 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008221_140 |
0.0748916194 |
- |
- |
PREDICTED: uncharacterized protein LOC105644226 [Jatropha curcas] |
| 13 |
Hb_006031_020 |
0.0755596817 |
- |
- |
PREDICTED: protein FLX-like 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006400_030 |
0.0774852469 |
- |
- |
hypothetical protein OsI_01237 [Oryza sativa Indica Group] |
| 15 |
Hb_058999_020 |
0.0780010189 |
- |
- |
PREDICTED: mitochondrial outer membrane import complex protein METAXIN [Jatropha curcas] |
| 16 |
Hb_005463_040 |
0.0787342081 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 17 |
Hb_005618_170 |
0.0792400903 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 18 |
Hb_001226_160 |
0.0799847575 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein B''-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002214_040 |
0.0802720283 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_025240_040 |
0.0806086463 |
- |
- |
hypothetical protein JCGZ_02438 [Jatropha curcas] |