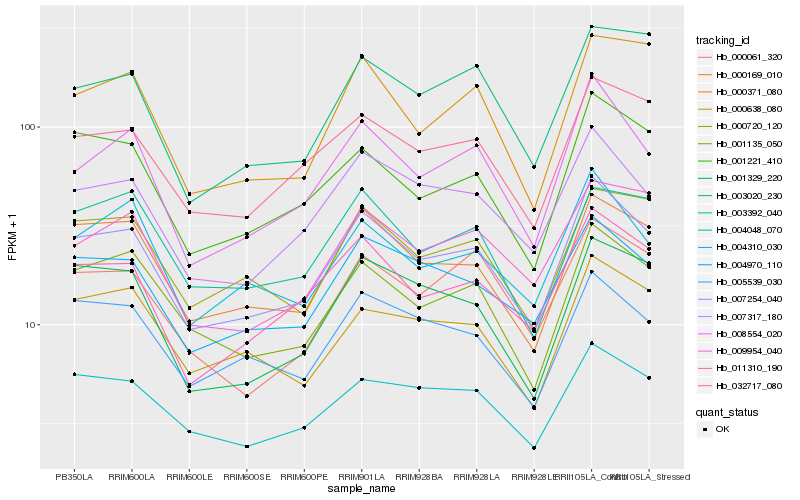
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007254_040 |
0.0 |
- |
- |
PREDICTED: protein MITOFERRINLIKE 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_008554_020 |
0.0611388044 |
- |
- |
- |
| 3 |
Hb_007317_180 |
0.0616339304 |
- |
- |
hypothetical protein CICLE_v10028933mg [Citrus clementina] |
| 4 |
Hb_000169_010 |
0.0620262959 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_011310_190 |
0.0631102962 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004970_110 |
0.064877986 |
- |
- |
PREDICTED: protein ROOT INITIATION DEFECTIVE 3 [Jatropha curcas] |
| 7 |
Hb_001135_050 |
0.0676294507 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_032717_080 |
0.0683178061 |
- |
- |
gamma-soluble nsf attachment protein, putative [Ricinus communis] |
| 9 |
Hb_004048_070 |
0.0700924504 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 6 [Jatropha curcas] |
| 10 |
Hb_001221_410 |
0.0735768387 |
- |
- |
PREDICTED: gamma carbonic anhydrase-like 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_005539_030 |
0.0749588753 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 12 |
Hb_004310_030 |
0.0759571846 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 13 |
Hb_001329_220 |
0.0789163576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000720_120 |
0.0814775147 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |
| 15 |
Hb_000638_080 |
0.0834890901 |
- |
- |
PREDICTED: uncharacterized protein LOC105632765 [Jatropha curcas] |
| 16 |
Hb_009954_040 |
0.0862350903 |
- |
- |
PREDICTED: putative golgin subfamily A member 6-like protein 6 [Jatropha curcas] |
| 17 |
Hb_000061_320 |
0.0881353456 |
- |
- |
PREDICTED: uncharacterized protein LOC105640575 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003392_040 |
0.0883578604 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Jatropha curcas] |
| 19 |
Hb_000371_080 |
0.0905012485 |
- |
- |
ubiquitin-conjugating enzyme m, putative [Ricinus communis] |
| 20 |
Hb_003020_230 |
0.0906402243 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |