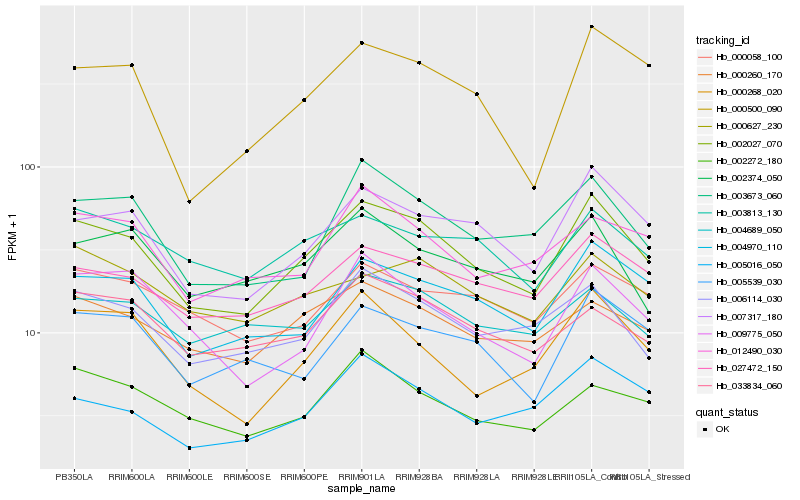
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_070 |
0.0 |
- |
- |
Auxin-binding protein T85 precursor, putative [Ricinus communis] |
| 2 |
Hb_007317_180 |
0.0817436663 |
- |
- |
hypothetical protein CICLE_v10028933mg [Citrus clementina] |
| 3 |
Hb_004970_110 |
0.0877610162 |
- |
- |
PREDICTED: protein ROOT INITIATION DEFECTIVE 3 [Jatropha curcas] |
| 4 |
Hb_027472_150 |
0.093721216 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 10 [Jatropha curcas] |
| 5 |
Hb_006114_030 |
0.0970812355 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 6 |
Hb_003673_060 |
0.0999599288 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 7 |
Hb_000500_090 |
0.1038284691 |
- |
- |
nucleoside diphosphate kinase 2 [Hevea brasiliensis] |
| 8 |
Hb_000260_170 |
0.1044974221 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 9 |
Hb_000268_020 |
0.1061446232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005016_050 |
0.1068781989 |
- |
- |
- |
| 11 |
Hb_005539_030 |
0.1083365099 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 12 |
Hb_009775_050 |
0.1087109133 |
transcription factor |
TF Family: Orphans |
hypothetical protein RCOM_0575190 [Ricinus communis] |
| 13 |
Hb_012490_030 |
0.1102532787 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_033834_060 |
0.1122710791 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_002374_050 |
0.1127851122 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 16 |
Hb_002272_180 |
0.1135202272 |
- |
- |
hypothetical protein JCGZ_23380 [Jatropha curcas] |
| 17 |
Hb_003813_130 |
0.1135333784 |
- |
- |
PREDICTED: plastidic glucose transporter 4 [Jatropha curcas] |
| 18 |
Hb_000058_100 |
0.1137567172 |
- |
- |
PREDICTED: RNA-directed DNA methylation 4 [Jatropha curcas] |
| 19 |
Hb_004689_050 |
0.1147250591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000627_230 |
0.1156171148 |
- |
- |
Uracil phosphoribosyltransferase, putative [Ricinus communis] |